Yixuan Shen
Criticality-Based Dynamic Topology Optimization for Enhancing Aerial-Marine Swarm Resilience
Aug 01, 2025Abstract:Heterogeneous marine-aerial swarm networks encounter substantial difficulties due to targeted communication disruptions and structural weaknesses in adversarial environments. This paper proposes a two-step framework to strengthen the network's resilience. Specifically, our framework combines the node prioritization based on criticality with multi-objective topology optimization. First, we design a three-layer architecture to represent structural, communication, and task dependencies of the swarm networks. Then, we introduce the SurBi-Ranking method, which utilizes graph convolutional networks, to dynamically evaluate and rank the criticality of nodes and edges in real time. Next, we apply the NSGA-III algorithm to optimize the network topology, aiming to balance communication efficiency, global connectivity, and mission success rate. Experiments demonstrate that compared to traditional methods like K-Shell, our SurBi-Ranking method identifies critical nodes and edges with greater accuracy, as deliberate attacks on these components cause more significant connectivity degradation. Furthermore, our optimization approach, when prioritizing SurBi-Ranked critical components under attack, reduces the natural connectivity degradation by around 30%, achieves higher mission success rates, and incurs lower communication reconfiguration costs, ensuring sustained connectivity and mission effectiveness across multi-phase operations.
From Histopathology Images to Cell Clouds: Learning Slide Representations with Hierarchical Cell Transformer
Dec 21, 2024Abstract:It is clinically crucial and potentially very beneficial to be able to analyze and model directly the spatial distributions of cells in histopathology whole slide images (WSI). However, most existing WSI datasets lack cell-level annotations, owing to the extremely high cost over giga-pixel images. Thus, it remains an open question whether deep learning models can directly and effectively analyze WSIs from the semantic aspect of cell distributions. In this work, we construct a large-scale WSI dataset with more than 5 billion cell-level annotations, termed WSI-Cell5B, and a novel hierarchical Cell Cloud Transformer (CCFormer) to tackle these challenges. WSI-Cell5B is based on 6,998 WSIs of 11 cancers from The Cancer Genome Atlas Program, and all WSIs are annotated per cell by coordinates and types. To the best of our knowledge, WSI-Cell5B is the first WSI-level large-scale dataset integrating cell-level annotations. On the other hand, CCFormer formulates the collection of cells in each WSI as a cell cloud and models cell spatial distribution. Specifically, Neighboring Information Embedding (NIE) is proposed to characterize the distribution of cells within the neighborhood of each cell, and a novel Hierarchical Spatial Perception (HSP) module is proposed to learn the spatial relationship among cells in a bottom-up manner. The clinical analysis indicates that WSI-Cell5B can be used to design clinical evaluation metrics based on counting cells that effectively assess the survival risk of patients. Extensive experiments on survival prediction and cancer staging show that learning from cell spatial distribution alone can already achieve state-of-the-art (SOTA) performance, i.e., CCFormer strongly outperforms other competing methods.
From Pixels to Gigapixels: Bridging Local Inductive Bias and Long-Range Dependencies with Pixel-Mamba
Dec 21, 2024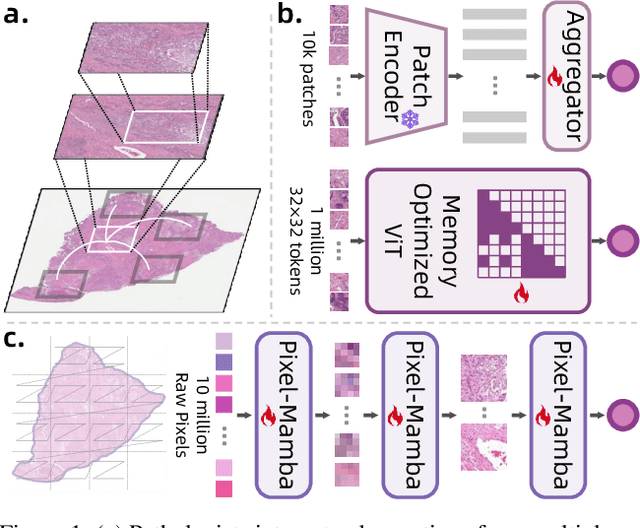
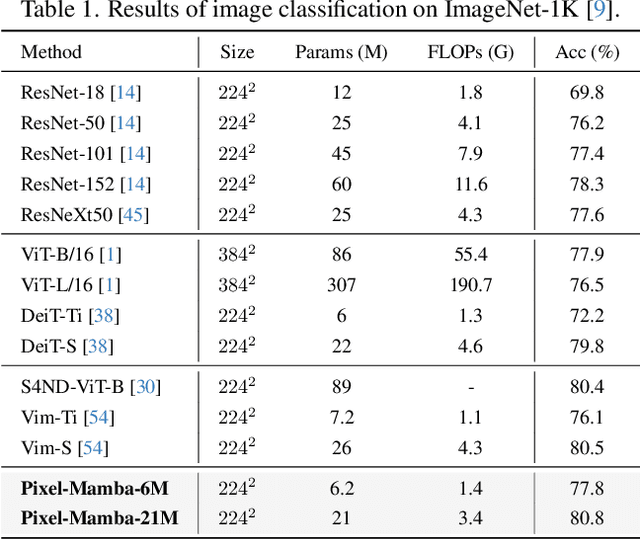
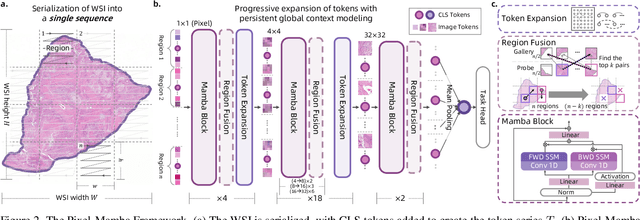
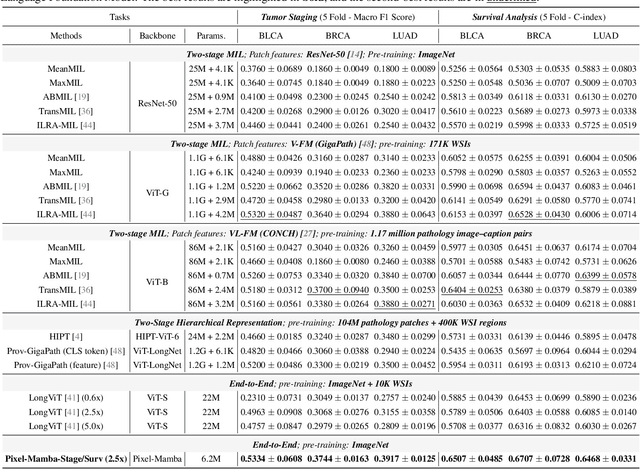
Abstract:Histopathology plays a critical role in medical diagnostics, with whole slide images (WSIs) offering valuable insights that directly influence clinical decision-making. However, the large size and complexity of WSIs may pose significant challenges for deep learning models, in both computational efficiency and effective representation learning. In this work, we introduce Pixel-Mamba, a novel deep learning architecture designed to efficiently handle gigapixel WSIs. Pixel-Mamba leverages the Mamba module, a state-space model (SSM) with linear memory complexity, and incorporates local inductive biases through progressively expanding tokens, akin to convolutional neural networks. This enables Pixel-Mamba to hierarchically combine both local and global information while efficiently addressing computational challenges. Remarkably, Pixel-Mamba achieves or even surpasses the quantitative performance of state-of-the-art (SOTA) foundation models that were pretrained on millions of WSIs or WSI-text pairs, in a range of tumor staging and survival analysis tasks, {\bf even without requiring any pathology-specific pretraining}. Extensive experiments demonstrate the efficacy of Pixel-Mamba as a powerful and efficient framework for end-to-end WSI analysis.
End-to-end Multi-source Visual Prompt Tuning for Survival Analysis in Whole Slide Images
Sep 05, 2024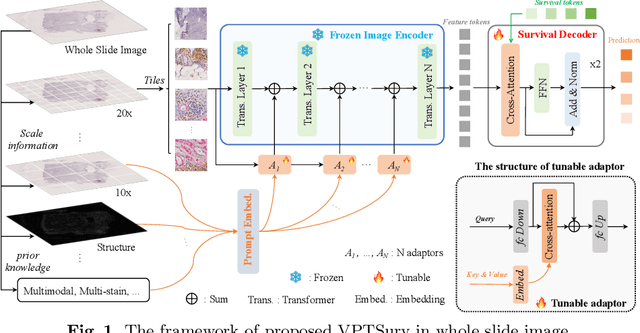
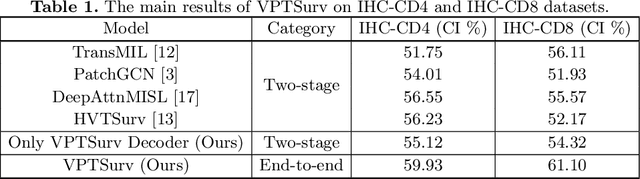
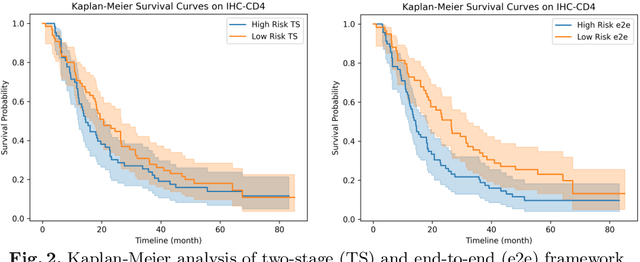
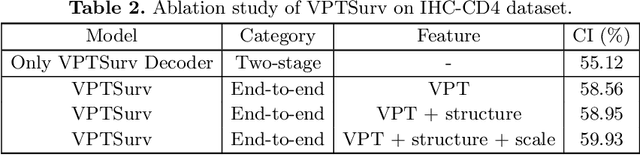
Abstract:Survival analysis using pathology images poses a considerable challenge, as it requires the localization of relevant information from the multitude of tiles within whole slide images (WSIs). Current methods typically resort to a two-stage approach, where a pre-trained network extracts features from tiles, which are then used by survival models. This process, however, does not optimize the survival models in an end-to-end manner, and the pre-extracted features may not be ideally suited for survival prediction. To address this limitation, we present a novel end-to-end Visual Prompt Tuning framework for survival analysis, named VPTSurv. VPTSurv refines feature embeddings through an efficient encoder-decoder framework. The encoder remains fixed while the framework introduces tunable visual prompts and adaptors, thus permitting end-to-end training specifically for survival prediction by optimizing only the lightweight adaptors and the decoder. Moreover, the versatile VPTSurv framework accommodates multi-source information as prompts, thereby enriching the survival model. VPTSurv achieves substantial increases of 8.7% and 12.5% in the C-index on two immunohistochemical pathology image datasets. These significant improvements highlight the transformative potential of the end-to-end VPT framework over traditional two-stage methods.
DIVESPOT: Depth Integrated Volume Estimation of Pile of Things Based on Point Cloud
Jul 07, 2024Abstract:Non-contact volume estimation of pile-type objects has considerable potential in industrial scenarios, including grain, coal, mining, and stone materials. However, using existing method for these scenarios is challenged by unstable measurement poses, significant light interference, the difficulty of training data collection, and the computational burden brought by large piles. To address the above issues, we propose the Depth Integrated Volume EStimation of Pile Of Things (DIVESPOT) based on point cloud technology in this study. For the challenges of unstable measurement poses, the point cloud pose correction and filtering algorithm is designed based on the Random Sample Consensus (RANSAC) and the Hierarchical Density-Based Spatial Clustering of Applications with Noise (HDBSCAN). To cope with light interference and to avoid the relying on training data, the height-distribution-based ground feature extraction algorithm is proposed to achieve RGB-independent. To reduce the computational burden, the storage space optimizing strategy is developed, such that accurate estimation can be acquired by using compressed voxels. Experimental results demonstrate that the DIVESPOT method enables non-data-driven, RGB-independent segmentation of pile point clouds, maintaining a volume calculation relative error within 2%. Even with 90% compression of the voxel mesh, the average error of the results can be under 3%.
Zero3D: Semantic-Driven Multi-Category 3D Shape Generation
Feb 13, 2023Abstract:Semantic-driven 3D shape generation aims to generate 3D objects conditioned on text. Previous works face problems with single-category generation, low-frequency 3D details, and requiring a large number of paired datasets for training. To tackle these challenges, we propose a multi-category conditional diffusion model. Specifically, 1) to alleviate the problem of lack of large-scale paired data, we bridge the text, 2D image and 3D shape based on the pre-trained CLIP model, and 2) to obtain the multi-category 3D shape feature, we apply the conditional flow model to generate 3D shape vector conditioned on CLIP embedding. 3) to generate multi-category 3D shape, we employ the hidden-layer diffusion model conditioned on the multi-category shape vector, which greatly reduces the training time and memory consumption.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge