Elliot K. Fishman
Learning Inductive Attention Guidance for Partially Supervised Pancreatic Ductal Adenocarcinoma Prediction
May 31, 2021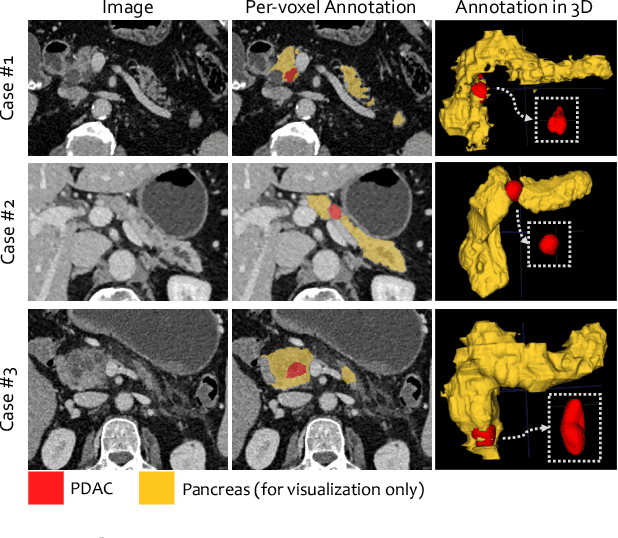
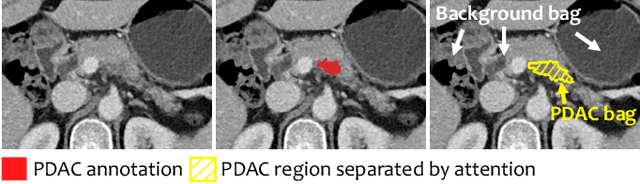
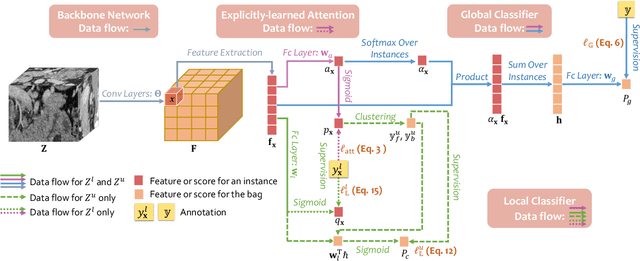
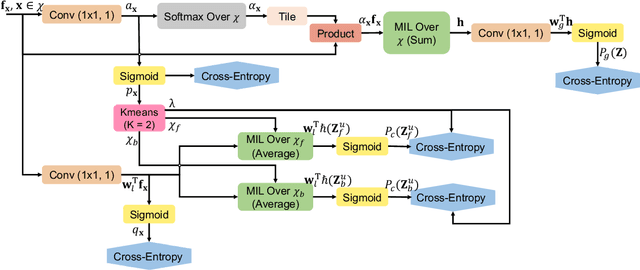
Abstract:Pancreatic ductal adenocarcinoma (PDAC) is the third most common cause of cancer death in the United States. Predicting tumors like PDACs (including both classification and segmentation) from medical images by deep learning is becoming a growing trend, but usually a large number of annotated data are required for training, which is very labor-intensive and time-consuming. In this paper, we consider a partially supervised setting, where cheap image-level annotations are provided for all the training data, and the costly per-voxel annotations are only available for a subset of them. We propose an Inductive Attention Guidance Network (IAG-Net) to jointly learn a global image-level classifier for normal/PDAC classification and a local voxel-level classifier for semi-supervised PDAC segmentation. We instantiate both the global and the local classifiers by multiple instance learning (MIL), where the attention guidance, indicating roughly where the PDAC regions are, is the key to bridging them: For global MIL based normal/PDAC classification, attention serves as a weight for each instance (voxel) during MIL pooling, which eliminates the distraction from the background; For local MIL based semi-supervised PDAC segmentation, the attention guidance is inductive, which not only provides bag-level pseudo-labels to training data without per-voxel annotations for MIL training, but also acts as a proxy of an instance-level classifier. Experimental results show that our IAG-Net boosts PDAC segmentation accuracy by more than 5% compared with the state-of-the-arts.
Multi-phase Deformable Registration for Time-dependent Abdominal Organ Variations
Mar 08, 2021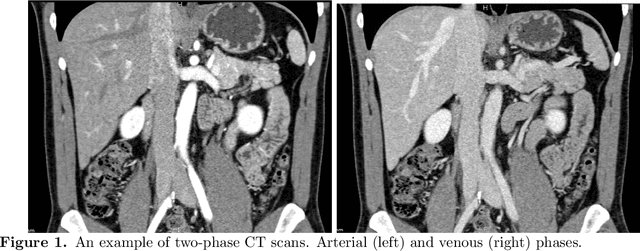
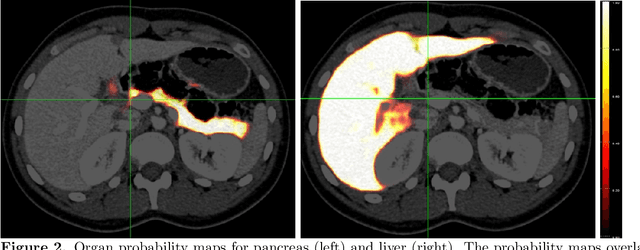

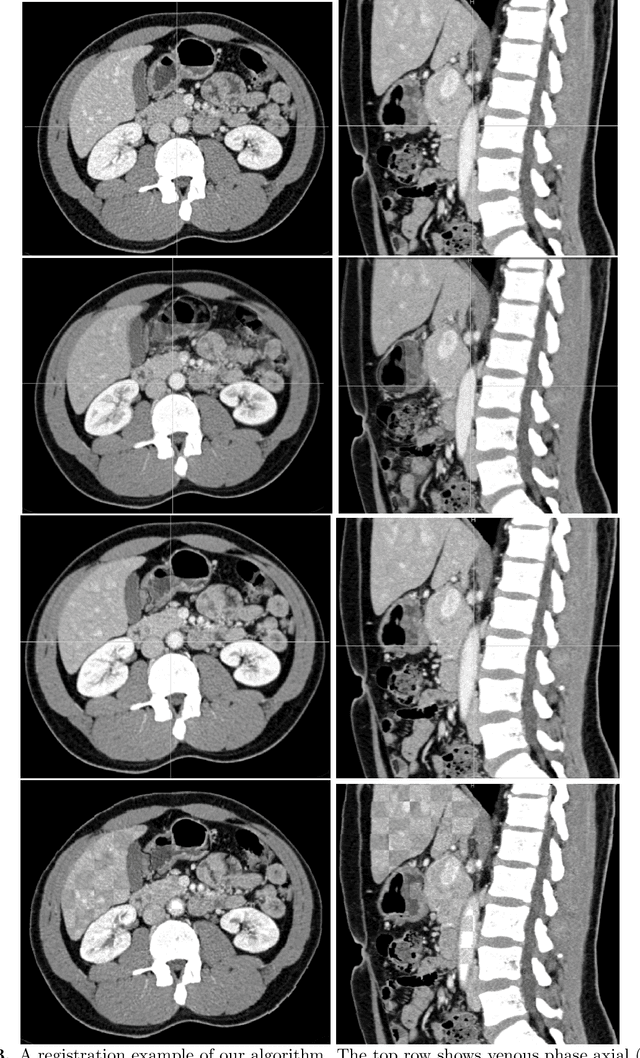
Abstract:Human body is a complex dynamic system composed of various sub-dynamic parts. Especially, thoracic and abdominal organs have complex internal shape variations with different frequencies by various reasons such as respiration with fast motion and peristalsis with slower motion. CT protocols for abdominal lesions are multi-phase scans for various tumor detection to use different vascular contrast, however, they are not aligned well enough to visually check the same area. In this paper, we propose a time-efficient and accurate deformable registration algorithm for multi-phase CT scans considering abdominal organ motions, which can be applied for differentiable or non-differentiable motions of abdominal organs. Experimental results shows the registration accuracy as 0.85 +/- 0.45mm (mean +/- STD) for pancreas within 1 minute for the whole abdominal region.
Volumetric Medical Image Segmentation: A 3D Deep Coarse-to-fine Framework and Its Adversarial Examples
Oct 29, 2020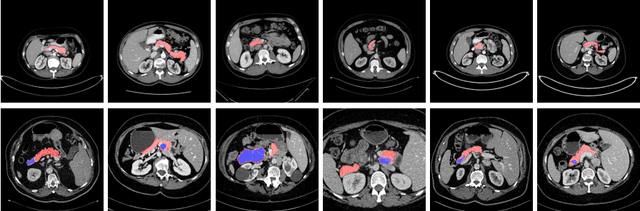
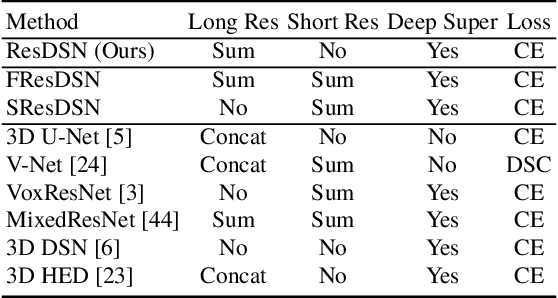
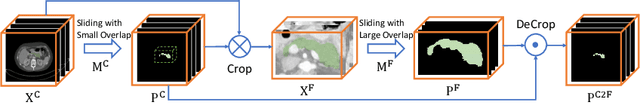
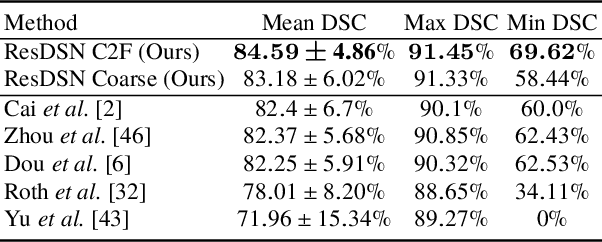
Abstract:Although deep neural networks have been a dominant method for many 2D vision tasks, it is still challenging to apply them to 3D tasks, such as medical image segmentation, due to the limited amount of annotated 3D data and limited computational resources. In this chapter, by rethinking the strategy to apply 3D Convolutional Neural Networks to segment medical images, we propose a novel 3D-based coarse-to-fine framework to efficiently tackle these challenges. The proposed 3D-based framework outperforms their 2D counterparts by a large margin since it can leverage the rich spatial information along all three axes. We further analyze the threat of adversarial attacks on the proposed framework and show how to defense against the attack. We conduct experiments on three datasets, the NIH pancreas dataset, the JHMI pancreas dataset and the JHMI pathological cyst dataset, where the first two and the last one contain healthy and pathological pancreases respectively, and achieve the current state-of-the-art in terms of Dice-Sorensen Coefficient (DSC) on all of them. Especially, on the NIH pancreas segmentation dataset, we outperform the previous best by an average of over $2\%$, and the worst case is improved by $7\%$ to reach almost $70\%$, which indicates the reliability of our framework in clinical applications.
Segmentation for Classification of Screening Pancreatic Neuroendocrine Tumors
Apr 04, 2020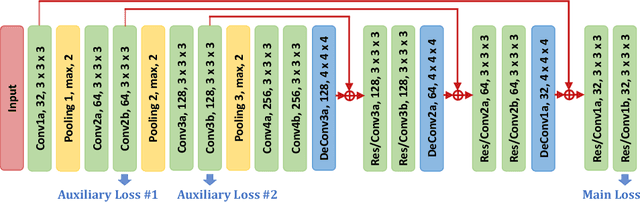
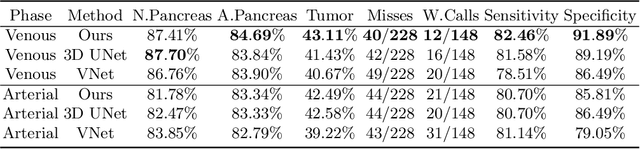
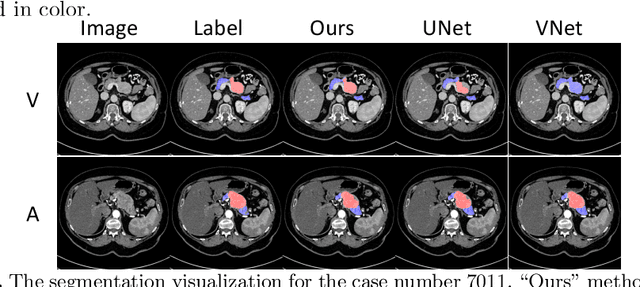
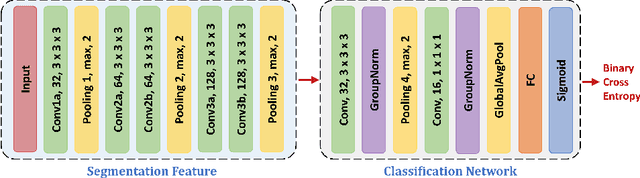
Abstract:This work presents comprehensive results to detect in the early stage the pancreatic neuroendocrine tumors (PNETs), a group of endocrine tumors arising in the pancreas, which are the second common type of pancreatic cancer, by checking the abdominal CT scans. To the best of our knowledge, this task has not been studied before as a computational task. To provide radiologists with tumor locations, we adopt a segmentation framework to classify CT volumes by checking if at least a sufficient number of voxels is segmented as tumors. To quantitatively analyze our method, we collect and voxelwisely label a new abdominal CT dataset containing $376$ cases with both arterial and venous phases available for each case, in which $228$ cases were diagnosed with PNETs while the remaining $148$ cases are normal, which is currently the largest dataset for PNETs to the best of our knowledge. In order to incorporate rich knowledge of radiologists to our framework, we annotate dilated pancreatic duct as well, which is regarded as the sign of high risk for pancreatic cancer. Quantitatively, our approach outperforms state-of-the-art segmentation networks and achieves a sensitivity of $89.47\%$ at a specificity of $81.08\%$, which indicates a potential direction to achieve a clinical impact related to cancer diagnosis by earlier tumor detection.
Detecting Pancreatic Adenocarcinoma in Multi-phase CT Scans via Alignment Ensemble
Apr 03, 2020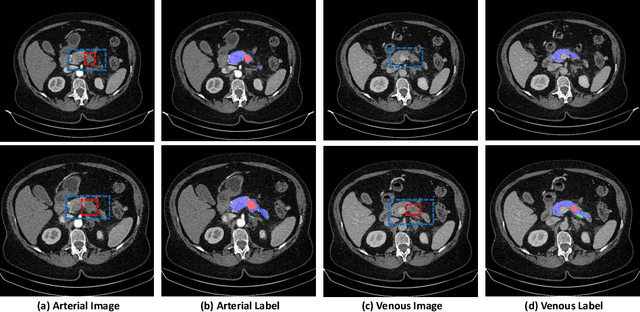

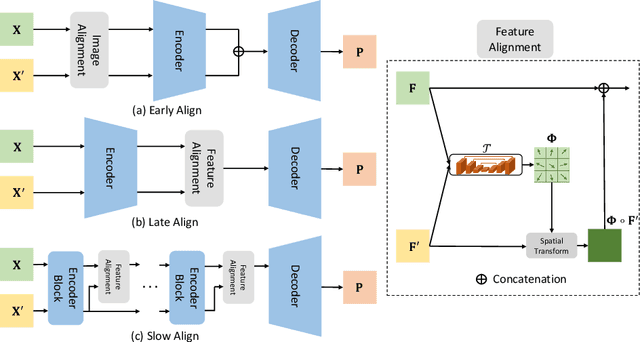

Abstract:Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal cancers among population. Screening for PDACs in dynamic contrast-enhanced CT is beneficial for early diagnose. In this paper, we investigate the problem of automated detecting PDACs in multi-phase (arterial and venous) CT scans. Multiple phases provide more information than single phase, but they are unaligned and inhomogeneous in texture, making it difficult to combine cross-phase information seamlessly. We study multiple phase alignment strategies, i.e., early alignment (image registration), late alignment (high-level feature registration) and slow alignment (multi-level feature registration), and suggest an ensemble of all these alignments as a promising way to boost the performance of PDAC detection. We provide an extensive empirical evaluation on two PDAC datasets and show that the proposed alignment ensemble significantly outperforms previous state-of-the-art approaches, illustrating strong potential for clinical use.
Deep Distance Transform for Tubular Structure Segmentation in CT Scans
Dec 06, 2019



Abstract:Tubular structure segmentation in medical images, e.g., segmenting vessels in CT scans, serves as a vital step in the use of computers to aid in screening early stages of related diseases. But automatic tubular structure segmentation in CT scans is a challenging problem, due to issues such as poor contrast, noise and complicated background. A tubular structure usually has a cylinder-like shape which can be well represented by its skeleton and cross-sectional radii (scales). Inspired by this, we propose a geometry-aware tubular structure segmentation method, Deep Distance Transform (DDT), which combines intuitions from the classical distance transform for skeletonization and modern deep segmentation networks. DDT first learns a multi-task network to predict a segmentation mask for a tubular structure and a distance map. Each value in the map represents the distance from each tubular structure voxel to the tubular structure surface. Then the segmentation mask is refined by leveraging the shape prior reconstructed from the distance map. We apply our DDT on six medical image datasets. The experiments show that (1) DDT can boost tubular structure segmentation performance significantly (e.g., over 13% improvement measured by DSC for pancreatic duct segmentation), and (2) DDT additionally provides a geometrical measurement for a tubular structure, which is important for clinical diagnosis (e.g., the cross-sectional scale of a pancreatic duct can be an indicator for pancreatic cancer).
Thickened 2D Networks for 3D Medical Image Segmentation
Apr 02, 2019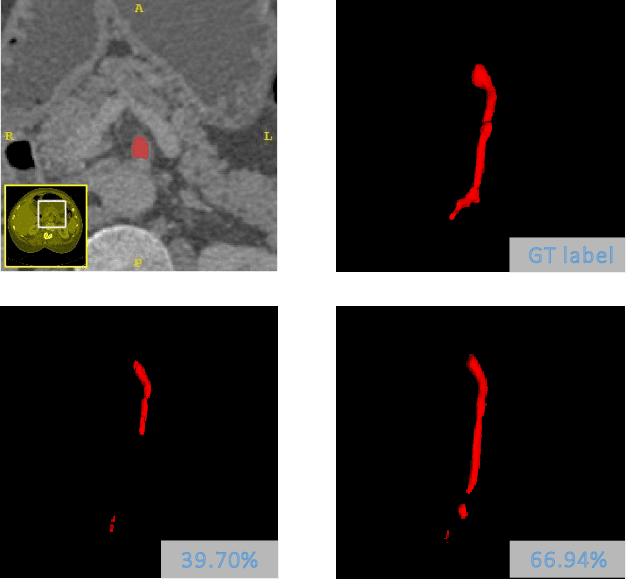
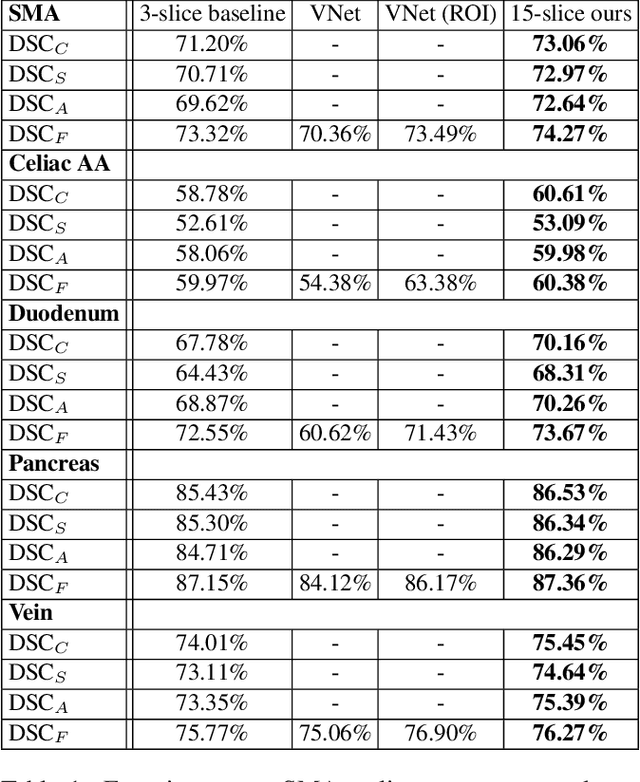
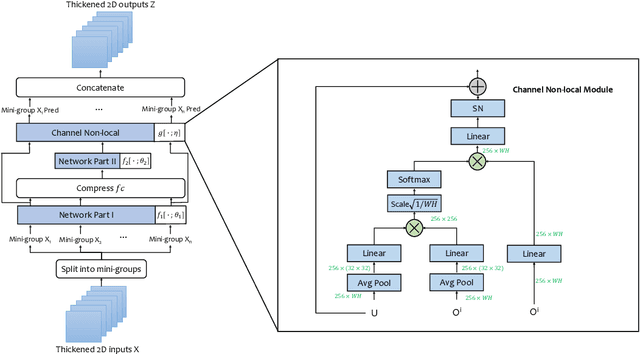
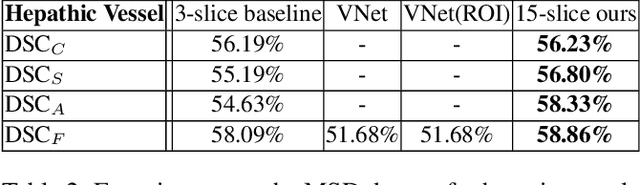
Abstract:There has been a debate in medical image segmentation on whether to use 2D or 3D networks, where both pipelines have advantages and disadvantages. This paper presents a novel approach which thickens the input of a 2D network, so that the model is expected to enjoy both the stability and efficiency of 2D networks as well as the ability of 3D networks in modeling volumetric contexts. A major information loss happens when a large number of 2D slices are fused at the first convolutional layer, resulting in a relatively weak ability of the network in distinguishing the difference among slices. To alleviate this drawback, we propose an effective framework which (i) postpones slice fusion and (ii) adds highway connections from the pre-fusion layer so that the prediction layer receives slice-sensitive auxiliary cues. Experiments on segmenting a few abdominal targets in particular blood vessels which require strong 3D contexts demonstrate the effectiveness of our approach.
Phase Collaborative Network for Multi-Phase Medical Imaging Segmentation
Dec 06, 2018



Abstract:Integrating multi-phase information is an effective way of boosting visual recognition. In this paper, we investigate this problem from the perspective of medical imaging analysis, in which two phases in CT scans known as arterial and venous are combined towards higher segmentation accuracy. To this end, we propose Phase Collaborative Network (PCN), an end-to-end network which contains both generative and discriminative modules to formulate phase-to-phase relations and data-to-label relations, respectively. Experiments are performed on several CT image segmentation datasets. PCN achieves superior performance with either two phases or only one phase available. Moreover, we empirically verify that the accuracy gain comes from the collaboration between phases.
Elastic Boundary Projection for 3D Medical Imaging Segmentation
Dec 03, 2018



Abstract:We focus on an important yet challenging problem: using a 2D deep network to deal with 3D segmentation for medical imaging analysis. Existing approaches either applied multi-view planar (2D) networks or directly used volumetric (3D) networks for this purpose, but both of them are not ideal: 2D networks cannot capture 3D contexts effectively, and 3D networks are both memory-consuming and less stable arguably due to the lack of pre-trained models. In this paper, we bridge the gap between 2D and 3D using a novel approach named Elastic Boundary Projection (EBP). The key observation is that, although the object is a 3D volume, what we really need in segmentation is to find its boundary which is a 2D surface. Therefore, we place a number of pivot points in the 3D space, and for each pivot, we determine its distance to the object boundary along a dense set of directions. This creates an elastic shell around each pivot which is initialized as a perfect sphere. We train a 2D deep network to determine whether each ending point falls within the object, and gradually adjust the shell so that it gradually converges to the actual shape of the boundary and thus achieves the goal of segmentation. EBP allows 3D segmentation without cutting the volume into slices or small patches, which stands out from conventional 2D and 3D approaches. EBP achieves promising accuracy in segmenting several abdominal organs from CT scans.
A 3D Coarse-to-Fine Framework for Volumetric Medical Image Segmentation
Aug 02, 2018



Abstract:In this paper, we adopt 3D Convolutional Neural Networks to segment volumetric medical images. Although deep neural networks have been proven to be very effective on many 2D vision tasks, it is still challenging to apply them to 3D tasks due to the limited amount of annotated 3D data and limited computational resources. We propose a novel 3D-based coarse-to-fine framework to effectively and efficiently tackle these challenges. The proposed 3D-based framework outperforms the 2D counterpart to a large margin since it can leverage the rich spatial infor- mation along all three axes. We conduct experiments on two datasets which include healthy and pathological pancreases respectively, and achieve the current state-of-the-art in terms of Dice-S{\o}rensen Coefficient (DSC). On the NIH pancreas segmentation dataset, we outperform the previous best by an average of over 2%, and the worst case is improved by 7% to reach almost 70%, which indicates the reliability of our framework in clinical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge