Seyoun Park
A Light-weight Interpretable CompositionalNetwork for Nuclei Detection and Weakly-supervised Segmentation
Oct 26, 2021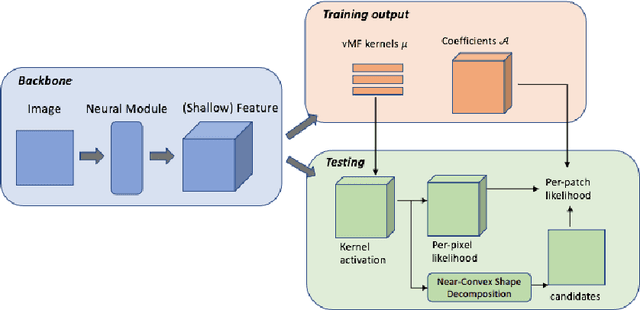

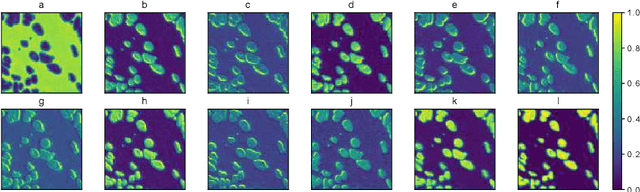
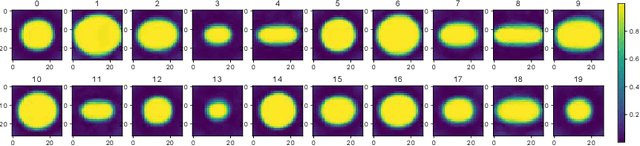
Abstract:The field of computational pathology has witnessed great advancements since deep neural networks have been widely applied. These deep neural networks usually require large numbers of annotated data to train vast parameters. However, it takes significant effort to annotate a large histopathology dataset. We propose to build a data-efficient model, which only requires partial annotation, specifically on isolated nucleus, rather than on the whole slide image. It exploits shallow features as its backbone and is light-weight, therefore a small number of data is sufficient for training. What's more, it is a generative compositional model, which enjoys interpretability in its prediction. The proposed method could be an alternative solution for the data-hungry problem of deep learning methods.
Multi-phase Deformable Registration for Time-dependent Abdominal Organ Variations
Mar 08, 2021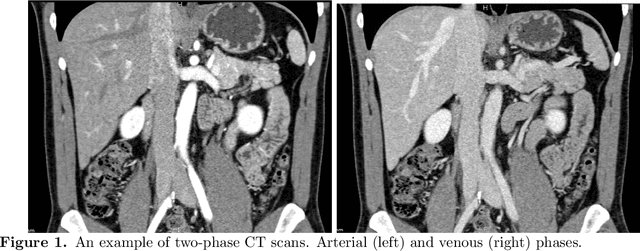
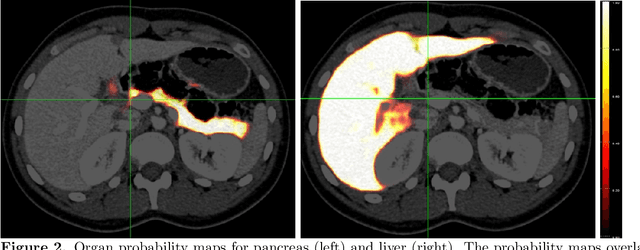

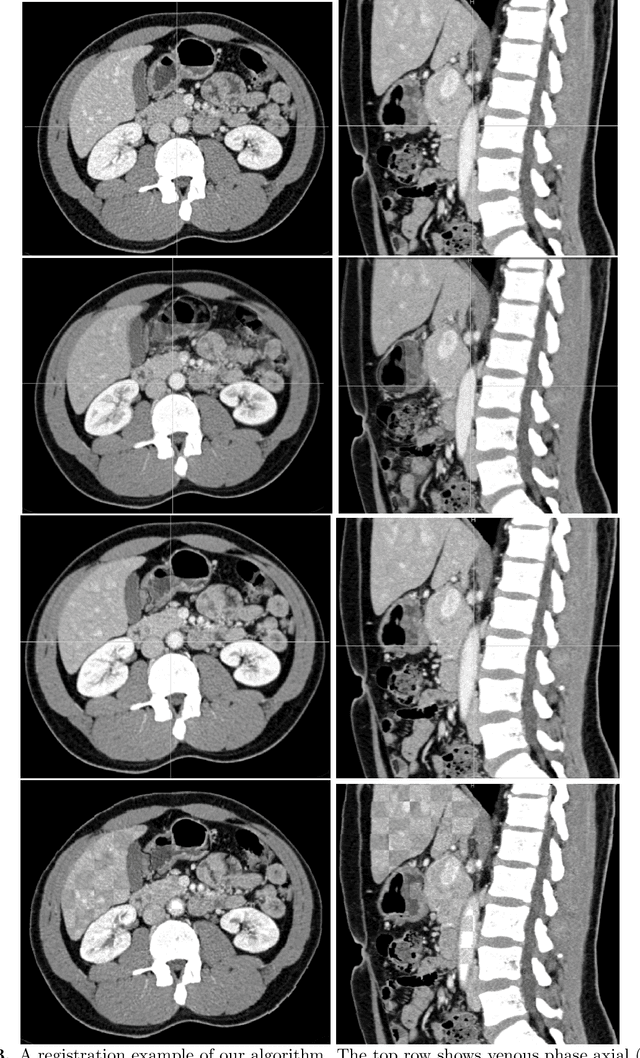
Abstract:Human body is a complex dynamic system composed of various sub-dynamic parts. Especially, thoracic and abdominal organs have complex internal shape variations with different frequencies by various reasons such as respiration with fast motion and peristalsis with slower motion. CT protocols for abdominal lesions are multi-phase scans for various tumor detection to use different vascular contrast, however, they are not aligned well enough to visually check the same area. In this paper, we propose a time-efficient and accurate deformable registration algorithm for multi-phase CT scans considering abdominal organ motions, which can be applied for differentiable or non-differentiable motions of abdominal organs. Experimental results shows the registration accuracy as 0.85 +/- 0.45mm (mean +/- STD) for pancreas within 1 minute for the whole abdominal region.
Hyper-Pairing Network for Multi-Phase Pancreatic Ductal Adenocarcinoma Segmentation
Sep 03, 2019



Abstract:Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal cancers with an overall five-year survival rate of 8%. Due to subtle texture changes of PDAC, pancreatic dual-phase imaging is recommended for better diagnosis of pancreatic disease. In this study, we aim at enhancing PDAC automatic segmentation by integrating multi-phase information (i.e., arterial phase and venous phase). To this end, we present Hyper-Pairing Network (HPN), a 3D fully convolution neural network which effectively integrates information from different phases. The proposed approach consists of a dual path network where the two parallel streams are interconnected with hyper-connections for intensive information exchange. Additionally, a pairing loss is added to encourage the commonality between high-level feature representations of different phases. Compared to prior arts which use single phase data, HPN reports a significant improvement up to 7.73% (from 56.21% to 63.94%) in terms of DSC.
Abdominal multi-organ segmentation with organ-attention networks and statistical fusion
Apr 23, 2018



Abstract:Accurate and robust segmentation of abdominal organs on CT is essential for many clinical applications such as computer-aided diagnosis and computer-aided surgery. But this task is challenging due to the weak boundaries of organs, the complexity of the background, and the variable sizes of different organs. To address these challenges, we introduce a novel framework for multi-organ segmentation by using organ-attention networks with reverse connections (OAN-RCs) which are applied to 2D views, of the 3D CT volume, and output estimates which are combined by statistical fusion exploiting structural similarity. OAN is a two-stage deep convolutional network, where deep network features from the first stage are combined with the original image, in a second stage, to reduce the complex background and enhance the discriminative information for the target organs. RCs are added to the first stage to give the lower layers semantic information thereby enabling them to adapt to the sizes of different organs. Our networks are trained on 2D views enabling us to use holistic information and allowing efficient computation. To compensate for the limited cross-sectional information of the original 3D volumetric CT, multi-sectional images are reconstructed from the three different 2D view directions. Then we combine the segmentation results from the different views using statistical fusion, with a novel term relating the structural similarity of the 2D views to the original 3D structure. To train the network and evaluate results, 13 structures were manually annotated by four human raters and confirmed by a senior expert on 236 normal cases. We tested our algorithm and computed Dice-Sorensen similarity coefficients and surface distances for evaluating our estimates of the 13 structures. Our experiments show that the proposed approach outperforms 2D- and 3D-patch based state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge