Janis Taube
A Light-weight Interpretable CompositionalNetwork for Nuclei Detection and Weakly-supervised Segmentation
Oct 26, 2021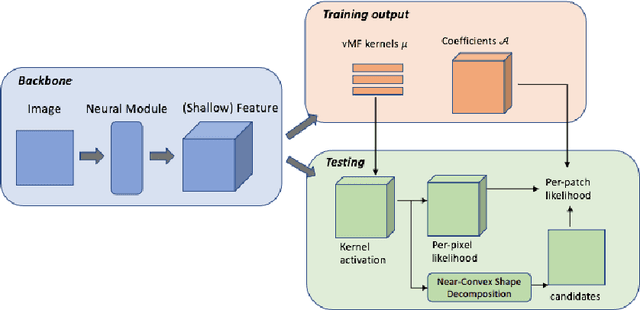

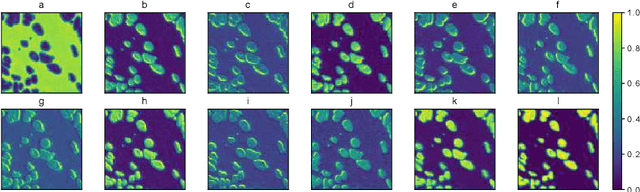
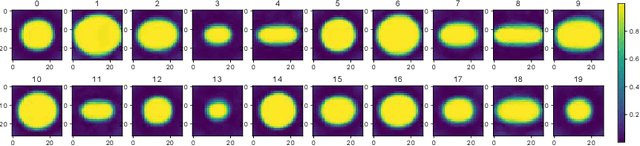
Abstract:The field of computational pathology has witnessed great advancements since deep neural networks have been widely applied. These deep neural networks usually require large numbers of annotated data to train vast parameters. However, it takes significant effort to annotate a large histopathology dataset. We propose to build a data-efficient model, which only requires partial annotation, specifically on isolated nucleus, rather than on the whole slide image. It exploits shallow features as its backbone and is light-weight, therefore a small number of data is sufficient for training. What's more, it is a generative compositional model, which enjoys interpretability in its prediction. The proposed method could be an alternative solution for the data-hungry problem of deep learning methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge