Elliot Fishman
Domain Adaptive Relational Reasoning for 3D Multi-Organ Segmentation
May 18, 2020



Abstract:In this paper, we present a novel unsupervised domain adaptation (UDA) method, named Domain Adaptive Relational Reasoning (DARR), to generalize 3D multi-organ segmentation models to medical data collected from different scanners and/or protocols (domains). Our method is inspired by the fact that the spatial relationship between internal structures in medical images is relatively fixed, e.g., a spleen is always located at the tail of a pancreas, which serves as a latent variable to transfer the knowledge shared across multiple domains. We formulate the spatial relationship by solving a jigsaw puzzle task, i.e., recovering a CT scan from its shuffled patches, and jointly train it with the organ segmentation task. To guarantee the transferability of the learned spatial relationship to multiple domains, we additionally introduce two schemes: 1) Employing a super-resolution network also jointly trained with the segmentation model to standardize medical images from different domain to a certain spatial resolution; 2) Adapting the spatial relationship for a test image by test-time jigsaw puzzle training. Experimental results show that our method improves the performance by 29.60\% DSC on target datasets on average without using any data from the target domain during training.
Hyper-Pairing Network for Multi-Phase Pancreatic Ductal Adenocarcinoma Segmentation
Sep 03, 2019



Abstract:Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal cancers with an overall five-year survival rate of 8%. Due to subtle texture changes of PDAC, pancreatic dual-phase imaging is recommended for better diagnosis of pancreatic disease. In this study, we aim at enhancing PDAC automatic segmentation by integrating multi-phase information (i.e., arterial phase and venous phase). To this end, we present Hyper-Pairing Network (HPN), a 3D fully convolution neural network which effectively integrates information from different phases. The proposed approach consists of a dual path network where the two parallel streams are interconnected with hyper-connections for intensive information exchange. Additionally, a pairing loss is added to encourage the commonality between high-level feature representations of different phases. Compared to prior arts which use single phase data, HPN reports a significant improvement up to 7.73% (from 56.21% to 63.94%) in terms of DSC.
FusionNet: Incorporating Shape and Texture for Abnormality Detection in 3D Abdominal CT Scans
Aug 27, 2019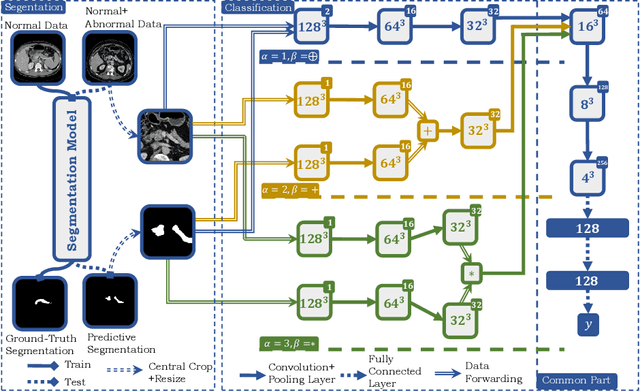



Abstract:Automatic abnormality detection in abdominal CT scans can help doctors improve the accuracy and efficiency in diagnosis. In this paper we aim at detecting pancreatic ductal adenocarcinoma (PDAC), the most common pancreatic cancer. Taking the fact that the existence of tumor can affect both the shape and the texture of pancreas, we design a system to extract the shape and texture feature at the same time for detecting PDAC. In this paper we propose a two-stage method for this 3D classification task. First, we segment the pancreas into a binary mask. Second, a FusionNet is proposed to take both the binary mask and CT image as input and perform a binary classification. The optimal architecture of the FusionNet is obtained by searching a pre-defined functional space. We show that the classification results using either shape or texture information are complementary, and by fusing them with the optimized architecture, the performance improves by a large margin. Our method achieves a specificity of 97% and a sensitivity of 92% on 200 normal scans and 136 scans with PDAC.
Prior-aware Neural Network for Partially-Supervised Multi-Organ Segmentation
Apr 12, 2019



Abstract:Accurate multi-organ abdominal CT segmentation is essential to many clinical applications such as computer-aided intervention. As data annotation requires massive human labor from experienced radiologists, it is common that training data are partially labeled, e.g., pancreas datasets only have the pancreas labeled while leaving the rest marked as background. However, these background labels can be misleading in multi-organ segmentation since the "background" usually contains some other organs of interest. To address the background ambiguity in these partially-labeled datasets, we propose Prior-aware Neural Network (PaNN) via explicitly incorporating anatomical priors on abdominal organ sizes, guiding the training process with domain-specific knowledge. More specifically, PaNN assumes that the average organ size distributions in the abdomen should approximate their empirical distributions, a prior statistics obtained from the fully-labeled dataset. As our training objective is difficult to be directly optimized using stochastic gradient descent [20], we propose to reformulate it in a min-max form and optimize it via the stochastic primal-dual gradient algorithm. PaNN achieves state-of-the-art performance on the MICCAI2015 challenge "Multi-Atlas Labeling Beyond the Cranial Vault", a competition on organ segmentation in the abdomen. We report an average Dice score of 84.97%, surpassing the prior art by a large margin of 3.27%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge