Yu-Dong Zhang
Deep Learning for Brain Age Estimation: A Systematic Review
Dec 07, 2022Abstract:Over the years, Machine Learning models have been successfully employed on neuroimaging data for accurately predicting brain age. Deviations from the healthy brain aging pattern are associated to the accelerated brain aging and brain abnormalities. Hence, efficient and accurate diagnosis techniques are required for eliciting accurate brain age estimations. Several contributions have been reported in the past for this purpose, resorting to different data-driven modeling methods. Recently, deep neural networks (also referred to as deep learning) have become prevalent in manifold neuroimaging studies, including brain age estimation. In this review, we offer a comprehensive analysis of the literature related to the adoption of deep learning for brain age estimation with neuroimaging data. We detail and analyze different deep learning architectures used for this application, pausing at research works published to date quantitatively exploring their application. We also examine different brain age estimation frameworks, comparatively exposing their advantages and weaknesses. Finally, the review concludes with an outlook towards future directions that should be followed by prospective studies. The ultimate goal of this paper is to establish a common and informed reference for newcomers and experienced researchers willing to approach brain age estimation by using deep learning models
Automatic Diagnosis of Myocarditis Disease in Cardiac MRI Modality using Deep Transformers and Explainable Artificial Intelligence
Oct 26, 2022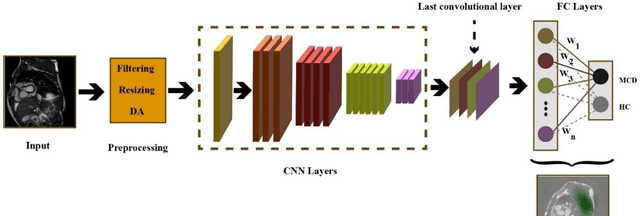
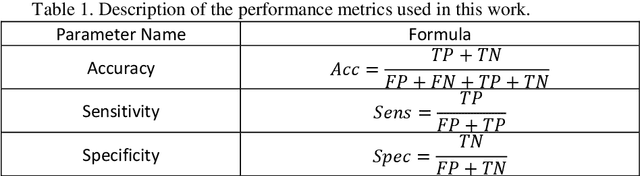
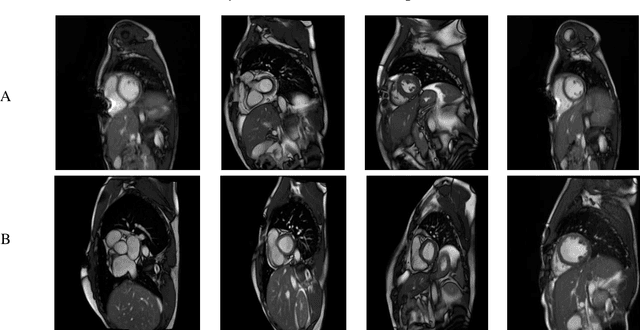
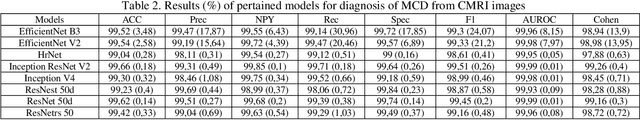
Abstract:Myocarditis is among the most important cardiovascular diseases (CVDs), endangering the health of many individuals by damaging the myocardium. Microbes and viruses, such as HIV, play a vital role in myocarditis disease (MCD) incidence. Lack of MCD diagnosis in the early stages is associated with irreversible complications. Cardiac magnetic resonance imaging (CMRI) is highly popular among cardiologists to diagnose CVDs. In this paper, a deep learning (DL) based computer-aided diagnosis system (CADS) is presented for the diagnosis of MCD using CMRI images. The proposed CADS includes dataset, preprocessing, feature extraction, classification, and post-processing steps. First, the Z-Alizadeh dataset was selected for the experiments. The preprocessing step included noise removal, image resizing, and data augmentation (DA). In this step, CutMix, and MixUp techniques were used for the DA. Then, the most recent pre-trained and transformers models were used for feature extraction and classification using CMRI images. Our results show high performance for the detection of MCD using transformer models compared with the pre-trained architectures. Among the DL architectures, Turbulence Neural Transformer (TNT) architecture achieved an accuracy of 99.73% with 10-fold cross-validation strategy. Explainable-based Grad Cam method is used to visualize the MCD suspected areas in CMRI images.
Automated Diagnosis of Cardiovascular Diseases from Cardiac Magnetic Resonance Imaging Using Deep Learning Models: A Review
Oct 26, 2022Abstract:In recent years, cardiovascular diseases (CVDs) have become one of the leading causes of mortality globally. CVDs appear with minor symptoms and progressively get worse. The majority of people experience symptoms such as exhaustion, shortness of breath, ankle swelling, fluid retention, and other symptoms when starting CVD. Coronary artery disease (CAD), arrhythmia, cardiomyopathy, congenital heart defect (CHD), mitral regurgitation, and angina are the most common CVDs. Clinical methods such as blood tests, electrocardiography (ECG) signals, and medical imaging are the most effective methods used for the detection of CVDs. Among the diagnostic methods, cardiac magnetic resonance imaging (CMR) is increasingly used to diagnose, monitor the disease, plan treatment and predict CVDs. Coupled with all the advantages of CMR data, CVDs diagnosis is challenging for physicians due to many slices of data, low contrast, etc. To address these issues, deep learning (DL) techniques have been employed to the diagnosis of CVDs using CMR data, and much research is currently being conducted in this field. This review provides an overview of the studies performed in CVDs detection using CMR images and DL techniques. The introduction section examined CVDs types, diagnostic methods, and the most important medical imaging techniques. In the following, investigations to detect CVDs using CMR images and the most significant DL methods are presented. Another section discussed the challenges in diagnosing CVDs from CMR data. Next, the discussion section discusses the results of this review, and future work in CVDs diagnosis from CMR images and DL techniques are outlined. The most important findings of this study are presented in the conclusion section.
Source-free unsupervised domain adaptation for cross-modality abdominal multi-organ segmentation
Nov 30, 2021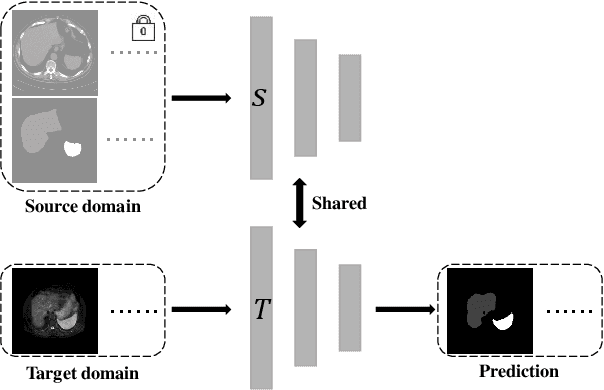
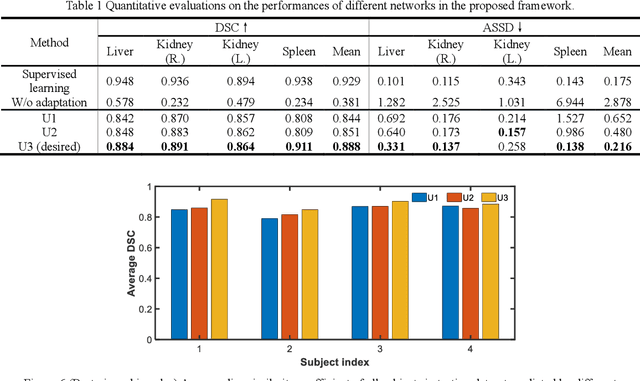
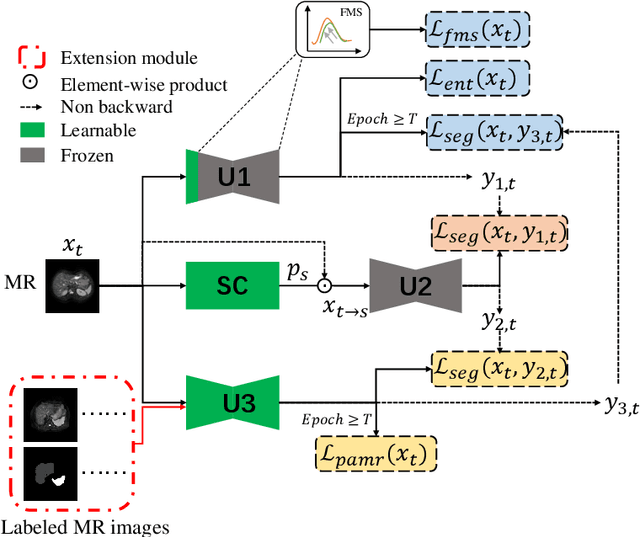
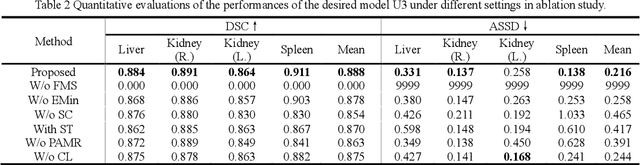
Abstract:It is valuable to achieve domain adaptation to transfer the learned knowledge from the source labeled CT dataset to the target unlabeled MR dataset for abdominal multi-organ segmentation. Meanwhile, it is highly desirable to avoid high annotation cost of target dataset and protect privacy of source dataset. Therefore, we propose an effective source-free unsupervised domain adaptation method for cross-modality abdominal multi-organ segmentation without accessing the source dataset. The process of the proposed framework includes two stages. At the first stage, the feature map statistics loss is used to align the distributions of the source and target features in the top segmentation network, and entropy minimization loss is used to encourage high confidence segmentations. The pseudo-labels outputted from the top segmentation network is used to guide the style compensation network to generate source-like images. The pseudo-labels outputted from the middle segmentation network is used to supervise the learning of the desired model (the bottom segmentation network). At the second stage, the circular learning and the pixel-adaptive mask refinement are used to further improve the performance of the desired model. With this approach, we achieve satisfactory performances on the segmentations of liver, right kidney, left kidney, and spleen with the dice similarity coefficients of 0.884, 0.891, 0.864, and 0.911, respectively. In addition, the proposed approach can be easily extended to the situation when there exists target annotation data. The performance improves from 0.888 to 0.922 in average dice similarity coefficient, close to the supervised learning (0.929), with only one labeled MR volume.
Sequential Learning on Liver Tumor Boundary Semantics and Prognostic Biomarker Mining
Mar 09, 2021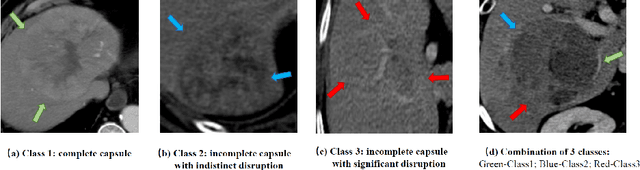
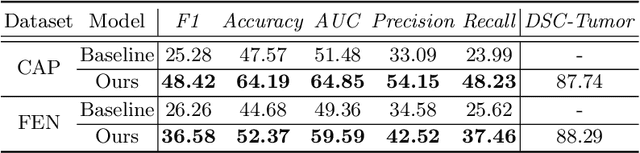
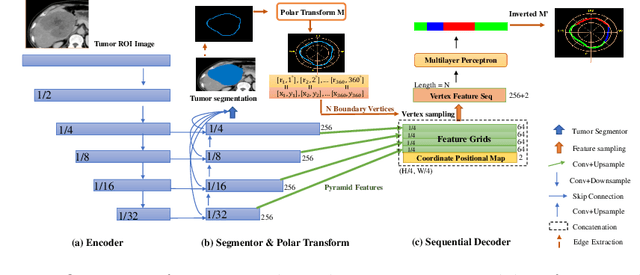
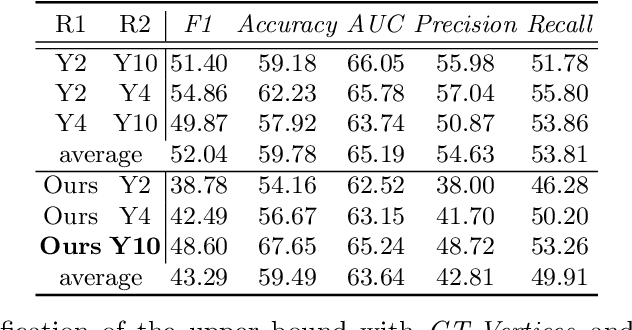
Abstract:The boundary of tumors (hepatocellular carcinoma, or HCC) contains rich semantics: capsular invasion, visibility, smoothness, folding and protuberance, etc. Capsular invasion on tumor boundary has proven to be clinically correlated with the prognostic indicator, microvascular invasion (MVI). Investigating tumor boundary semantics has tremendous clinical values. In this paper, we propose the first and novel computational framework that disentangles the task into two components: spatial vertex localization and sequential semantic classification. (1) A HCC tumor segmentor is built for tumor mask boundary extraction, followed by polar transform representing the boundary with radius and angle. Vertex generator is used to produce fixed-length boundary vertices where vertex features are sampled on the corresponding spatial locations. (2) The sampled deep vertex features with positional embedding are mapped into a sequential space and decoded by a multilayer perceptron (MLP) for semantic classification. Extensive experiments on tumor capsule semantics demonstrate the effectiveness of our framework. Mining the correlation between the boundary semantics and MVI status proves the feasibility to integrate this boundary semantics as a valid HCC prognostic biomarker.
Probabilistic combination of eigenlungs-based classifiers for COVID-19 diagnosis in chest CT images
Mar 04, 2021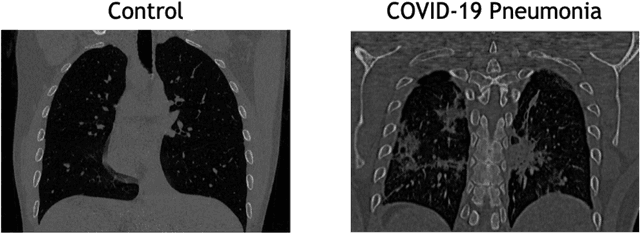
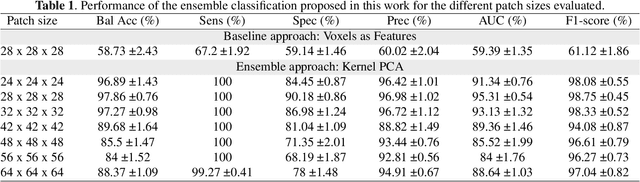
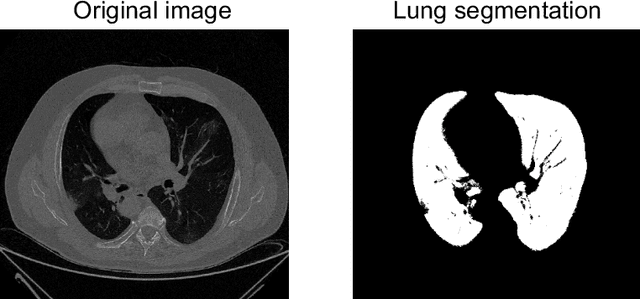
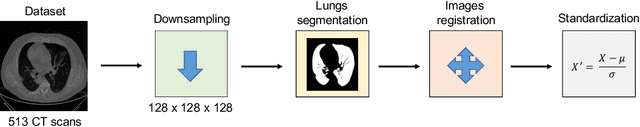
Abstract:The outbreak of the COVID-19 (Coronavirus disease 2019) pandemic has changed the world. According to the World Health Organization (WHO), there have been more than 100 million confirmed cases of COVID-19, including more than 2.4 million deaths. It is extremely important the early detection of the disease, and the use of medical imaging such as chest X-ray (CXR) and chest Computed Tomography (CCT) have proved to be an excellent solution. However, this process requires clinicians to do it within a manual and time-consuming task, which is not ideal when trying to speed up the diagnosis. In this work, we propose an ensemble classifier based on probabilistic Support Vector Machine (SVM) in order to identify pneumonia patterns while providing information about the reliability of the classification. Specifically, each CCT scan is divided into cubic patches and features contained in each one of them are extracted by applying kernel PCA. The use of base classifiers within an ensemble allows our system to identify the pneumonia patterns regardless of their size or location. Decisions of each individual patch are then combined into a global one according to the reliability of each individual classification: the lower the uncertainty, the higher the contribution. Performance is evaluated in a real scenario, yielding an accuracy of 97.86%. The large performance obtained and the simplicity of the system (use of deep learning in CCT images would result in a huge computational cost) evidence the applicability of our proposal in a real-world environment.
Uncertainty-driven ensembles of deep architectures for multiclass classification. Application to COVID-19 diagnosis in chest X-ray images
Nov 27, 2020
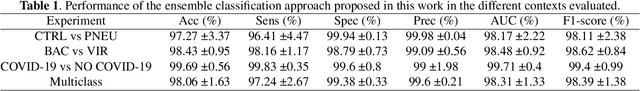
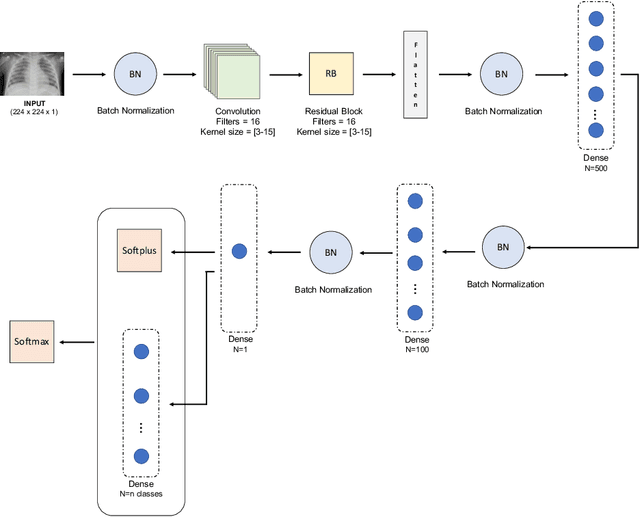
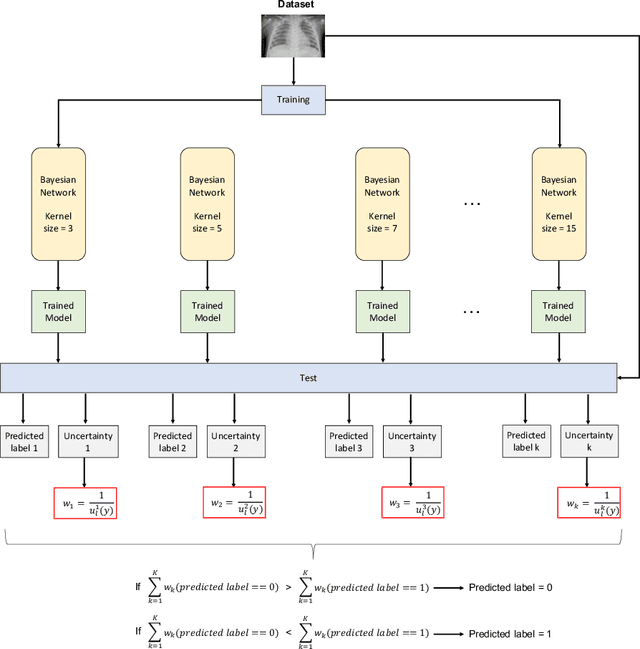
Abstract:Respiratory diseases kill million of people each year. Diagnosis of these pathologies is a manual, time-consuming process that has inter and intra-observer variability, delaying diagnosis and treatment. The recent COVID-19 pandemic has demonstrated the need of developing systems to automatize the diagnosis of pneumonia, whilst Convolutional Neural Network (CNNs) have proved to be an excellent option for the automatic classification of medical images. However, given the need of providing a confidence classification in this context it is crucial to quantify the reliability of the model's predictions. In this work, we propose a multi-level ensemble classification system based on a Bayesian Deep Learning approach in order to maximize performance while quantifying the uncertainty of each classification decision. This tool combines the information extracted from different architectures by weighting their results according to the uncertainty of their predictions. Performance of the Bayesian network is evaluated in a real scenario where simultaneously differentiating between four different pathologies: control vs bacterial pneumonia vs viral pneumonia vs COVID-19 pneumonia. A three-level decision tree is employed to divide the 4-class classification into three binary classifications, yielding an accuracy of 98.06% and overcoming the results obtained by recent literature. The reduced preprocessing needed for obtaining this high performance, in addition to the information provided about the reliability of the predictions evidence the applicability of the system to be used as an aid for clinicians.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge