Javier Ramírez
EEG Connectivity Analysis Using Denoising Autoencoders for the Detection of Dyslexia
Nov 23, 2023Abstract:The Temporal Sampling Framework (TSF) theorizes that the characteristic phonological difficulties of dyslexia are caused by an atypical oscillatory sampling at one or more temporal rates. The LEEDUCA study conducted a series of Electroencephalography (EEG) experiments on children listening to amplitude modulated (AM) noise with slow-rythmic prosodic (0.5-1 Hz), syllabic (4-8 Hz) or the phoneme (12-40 Hz) rates, aimed at detecting differences in perception of oscillatory sampling that could be associated with dyslexia. The purpose of this work is to check whether these differences exist and how they are related to children's performance in different language and cognitive tasks commonly used to detect dyslexia. To this purpose, temporal and spectral inter-channel EEG connectivity was estimated, and a denoising autoencoder (DAE) was trained to learn a low-dimensional representation of the connectivity matrices. This representation was studied via correlation and classification analysis, which revealed ability in detecting dyslexic subjects with an accuracy higher than 0.8, and balanced accuracy around 0.7. Some features of the DAE representation were significantly correlated ($p<0.005$) with children's performance in language and cognitive tasks of the phonological hypothesis category such as phonological awareness and rapid symbolic naming, as well as reading efficiency and reading comprehension. Finally, a deeper analysis of the adjacency matrix revealed a reduced bilateral connection between electrodes of the temporal lobe (roughly the primary auditory cortex) in DD subjects, as well as an increased connectivity of the F7 electrode, placed roughly on Broca's area. These results pave the way for a complementary assessment of dyslexia using more objective methodologies such as EEG.
* 19 pages, 6 figures
Convolutional Neural Networks for Neuroimaging in Parkinson's Disease: Is Preprocessing Needed?
Nov 21, 2023



Abstract:Spatial and intensity normalization are nowadays a prerequisite for neuroimaging analysis. Influenced by voxel-wise and other univariate comparisons, where these corrections are key, they are commonly applied to any type of analysis and imaging modalities. Nuclear imaging modalities such as PET-FDG or FP-CIT SPECT, a common modality used in Parkinson's Disease diagnosis, are especially dependent on intensity normalization. However, these steps are computationally expensive and furthermore, they may introduce deformations in the images, altering the information contained in them. Convolutional Neural Networks (CNNs), for their part, introduce position invariance to pattern recognition, and have been proven to classify objects regardless of their orientation, size, angle, etc. Therefore, a question arises: how well can CNNs account for spatial and intensity differences when analysing nuclear brain imaging? Are spatial and intensity normalization still needed? To answer this question, we have trained four different CNN models based on well-established architectures, using or not different spatial and intensity normalization preprocessing. The results show that a sufficiently complex model such as our three-dimensional version of the ALEXNET can effectively account for spatial differences, achieving a diagnosis accuracy of 94.1% with an area under the ROC curve of 0.984. The visualization of the differences via saliency maps shows that these models are correctly finding patterns that match those found in the literature, without the need of applying any complex spatial normalization procedure. However, the intensity normalization -- and its type -- is revealed as very influential in the results and accuracy of the trained model, and therefore must be well accounted.
* 19 pages, 7 figures
Tiled sparse coding in eigenspaces for the COVID-19 diagnosis in chest X-ray images
Jun 28, 2021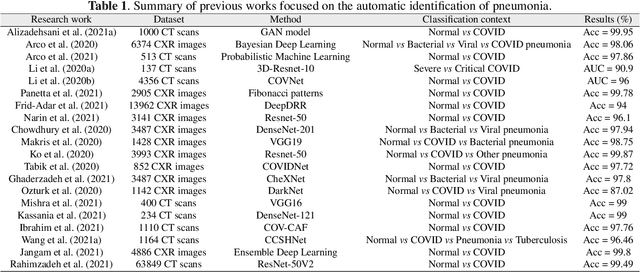
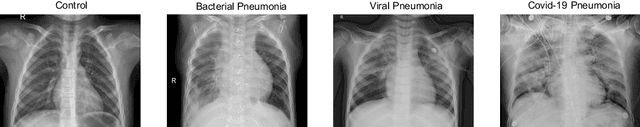
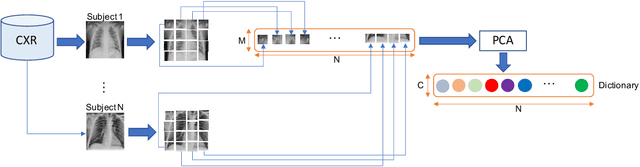
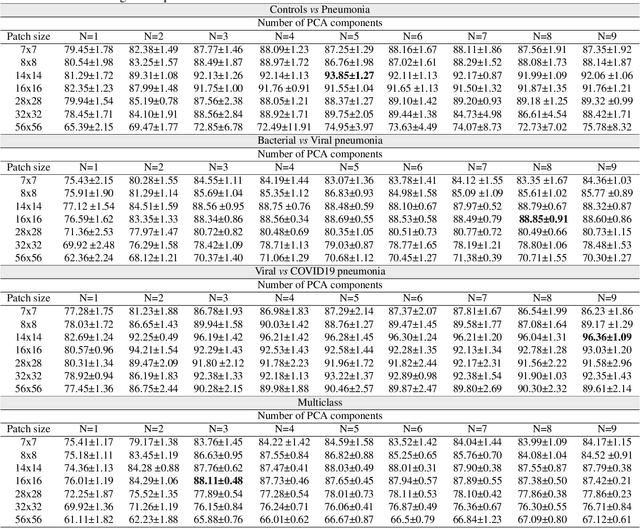
Abstract:The ongoing crisis of the COVID-19 (Coronavirus disease 2019) pandemic has changed the world. According to the World Health Organization (WHO), 4 million people have died due to this disease, whereas there have been more than 180 million confirmed cases of COVID-19. The collapse of the health system in many countries has demonstrated the need of developing tools to automatize the diagnosis of the disease from medical imaging. Previous studies have used deep learning for this purpose. However, the performance of this alternative highly depends on the size of the dataset employed for training the algorithm. In this work, we propose a classification framework based on sparse coding in order to identify the pneumonia patterns associated with different pathologies. Specifically, each chest X-ray (CXR) image is partitioned into different tiles. The most relevant features extracted from PCA are then used to build the dictionary within the sparse coding procedure. Once images are transformed and reconstructed from the elements of the dictionary, classification is performed from the reconstruction errors of individual patches associated with each image. Performance is evaluated in a real scenario where simultaneously differentiation between four different pathologies: control vs bacterial pneumonia vs viral pneumonia vs COVID-19. The accuracy when identifying the presence of pneumonia is 93.85%, whereas 88.11% is obtained in the 4-class classification context. The excellent results and the pioneering use of sparse coding in this scenario evidence the applicability of this approach as an aid for clinicians in a real-world environment.
Applications of Epileptic Seizures Detection in Neuroimaging Modalities Using Deep Learning Techniques: Methods, Challenges, and Future Works
May 29, 2021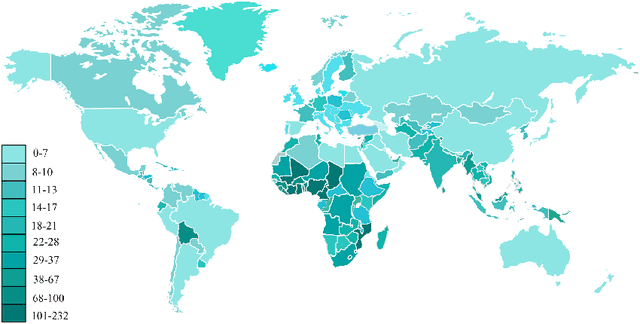
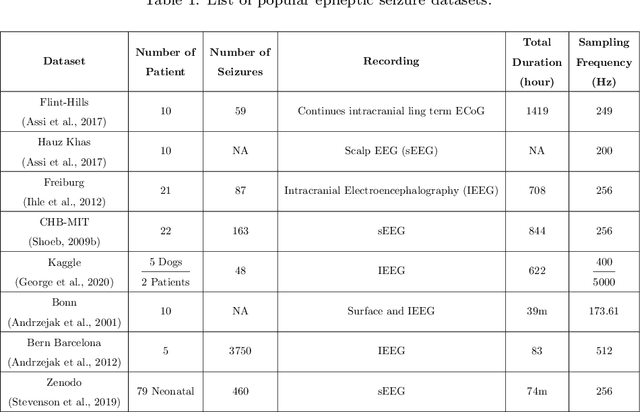
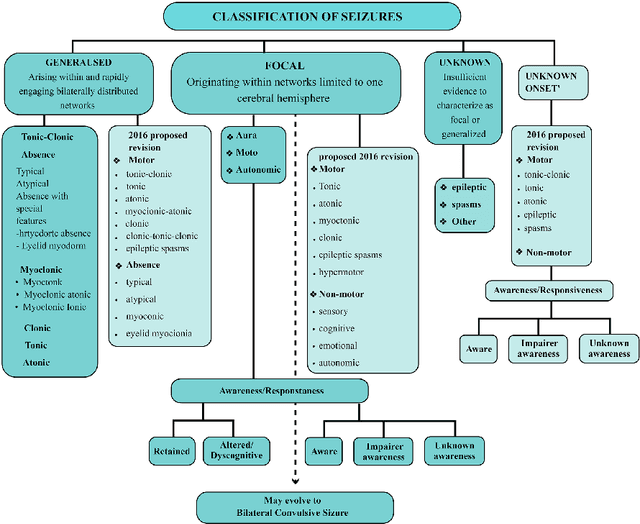
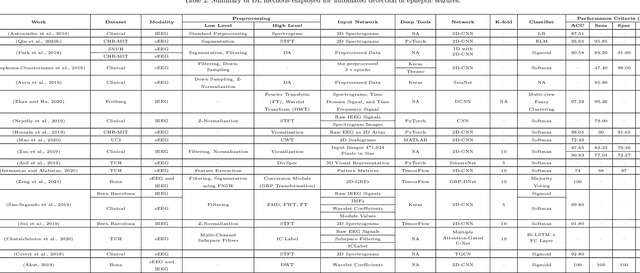
Abstract:Epileptic seizures are a type of neurological disorder that affect many people worldwide. Specialist physicians and neurologists take advantage of structural and functional neuroimaging modalities to diagnose various types of epileptic seizures. Neuroimaging modalities assist specialist physicians considerably in analyzing brain tissue and the changes made in it. One method to accelerate the accurate and fast diagnosis of epileptic seizures is to employ computer aided diagnosis systems (CADS) based on artificial intelligence (AI) and functional and structural neuroimaging modalities. AI encompasses a variety of areas, and one of its branches is deep learning (DL). Not long ago, and before the rise of DL algorithms, feature extraction was an essential part of every conventional machine learning method, yet handcrafting features limit these models' performances to the knowledge of system designers. DL methods resolved this issue entirely by automating the feature extraction and classification process; applications of these methods in many fields of medicine, such as the diagnosis of epileptic seizures, have made notable improvements. In this paper, a comprehensive overview of the types of DL methods exploited to diagnose epileptic seizures from various neuroimaging modalities has been studied. Additionally, rehabilitation systems and cloud computing in epileptic seizures diagnosis applications have been exactly investigated using various modalities.
Deep Learning in current Neuroimaging: a multivariate approach with power and type I error control but arguable generalization ability
Mar 30, 2021
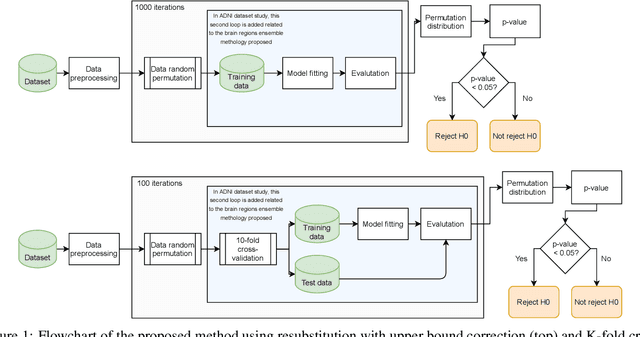
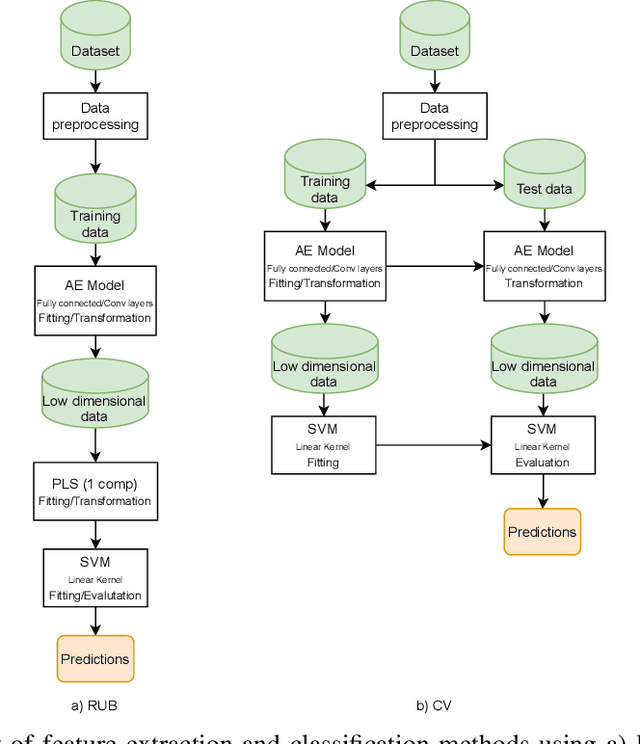
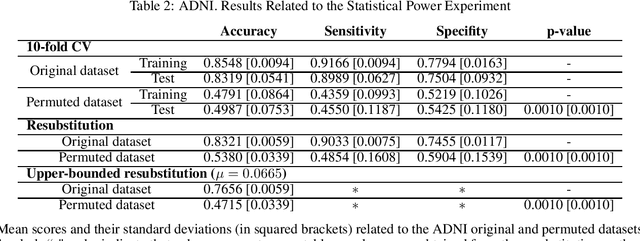
Abstract:Discriminative analysis in neuroimaging by means of deep/machine learning techniques is usually tested with validation techniques, whereas the associated statistical significance remains largely under-developed due to their computational complexity. In this work, a non-parametric framework is proposed that estimates the statistical significance of classifications using deep learning architectures. In particular, a combination of autoencoders (AE) and support vector machines (SVM) is applied to: (i) a one-condition, within-group designs often of normal controls (NC) and; (ii) a two-condition, between-group designs which contrast, for example, Alzheimer's disease (AD) patients with NC (the extension to multi-class analyses is also included). A random-effects inference based on a label permutation test is proposed in both studies using cross-validation (CV) and resubstitution with upper bound correction (RUB) as validation methods. This allows both false positives and classifier overfitting to be detected as well as estimating the statistical power of the test. Several experiments were carried out using the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset, the Dominantly Inherited Alzheimer Network (DIAN) dataset, and a MCI prediction dataset. We found in the permutation test that CV and RUB methods offer a false positive rate close to the significance level and an acceptable statistical power (although lower using cross-validation). A large separation between training and test accuracies using CV was observed, especially in one-condition designs. This implies a low generalization ability as the model fitted in training is not informative with respect to the test set. We propose as solution by applying RUB, whereby similar results are obtained to those of the CV test set, but considering the whole set and with a lower computational cost per iteration.
Probabilistic combination of eigenlungs-based classifiers for COVID-19 diagnosis in chest CT images
Mar 04, 2021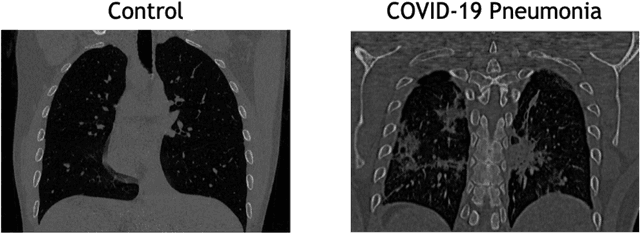
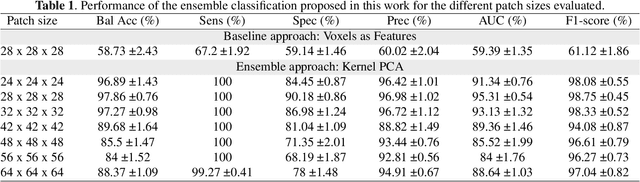
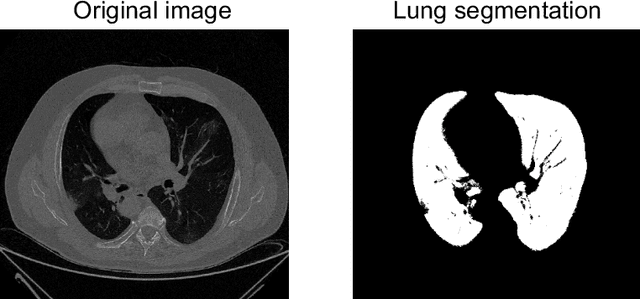
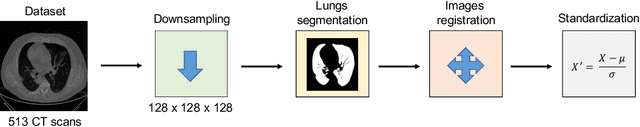
Abstract:The outbreak of the COVID-19 (Coronavirus disease 2019) pandemic has changed the world. According to the World Health Organization (WHO), there have been more than 100 million confirmed cases of COVID-19, including more than 2.4 million deaths. It is extremely important the early detection of the disease, and the use of medical imaging such as chest X-ray (CXR) and chest Computed Tomography (CCT) have proved to be an excellent solution. However, this process requires clinicians to do it within a manual and time-consuming task, which is not ideal when trying to speed up the diagnosis. In this work, we propose an ensemble classifier based on probabilistic Support Vector Machine (SVM) in order to identify pneumonia patterns while providing information about the reliability of the classification. Specifically, each CCT scan is divided into cubic patches and features contained in each one of them are extracted by applying kernel PCA. The use of base classifiers within an ensemble allows our system to identify the pneumonia patterns regardless of their size or location. Decisions of each individual patch are then combined into a global one according to the reliability of each individual classification: the lower the uncertainty, the higher the contribution. Performance is evaluated in a real scenario, yielding an accuracy of 97.86%. The large performance obtained and the simplicity of the system (use of deep learning in CCT images would result in a huge computational cost) evidence the applicability of our proposal in a real-world environment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge