Juan Manuel Górriz
EEG Connectivity Analysis Using Denoising Autoencoders for the Detection of Dyslexia
Nov 23, 2023Abstract:The Temporal Sampling Framework (TSF) theorizes that the characteristic phonological difficulties of dyslexia are caused by an atypical oscillatory sampling at one or more temporal rates. The LEEDUCA study conducted a series of Electroencephalography (EEG) experiments on children listening to amplitude modulated (AM) noise with slow-rythmic prosodic (0.5-1 Hz), syllabic (4-8 Hz) or the phoneme (12-40 Hz) rates, aimed at detecting differences in perception of oscillatory sampling that could be associated with dyslexia. The purpose of this work is to check whether these differences exist and how they are related to children's performance in different language and cognitive tasks commonly used to detect dyslexia. To this purpose, temporal and spectral inter-channel EEG connectivity was estimated, and a denoising autoencoder (DAE) was trained to learn a low-dimensional representation of the connectivity matrices. This representation was studied via correlation and classification analysis, which revealed ability in detecting dyslexic subjects with an accuracy higher than 0.8, and balanced accuracy around 0.7. Some features of the DAE representation were significantly correlated ($p<0.005$) with children's performance in language and cognitive tasks of the phonological hypothesis category such as phonological awareness and rapid symbolic naming, as well as reading efficiency and reading comprehension. Finally, a deeper analysis of the adjacency matrix revealed a reduced bilateral connection between electrodes of the temporal lobe (roughly the primary auditory cortex) in DD subjects, as well as an increased connectivity of the F7 electrode, placed roughly on Broca's area. These results pave the way for a complementary assessment of dyslexia using more objective methodologies such as EEG.
* 19 pages, 6 figures
Deep Learning in current Neuroimaging: a multivariate approach with power and type I error control but arguable generalization ability
Mar 30, 2021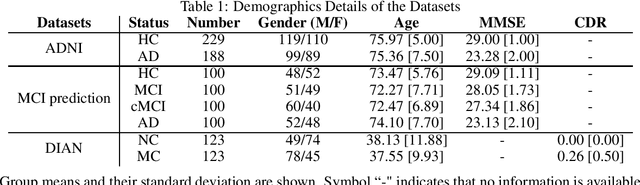
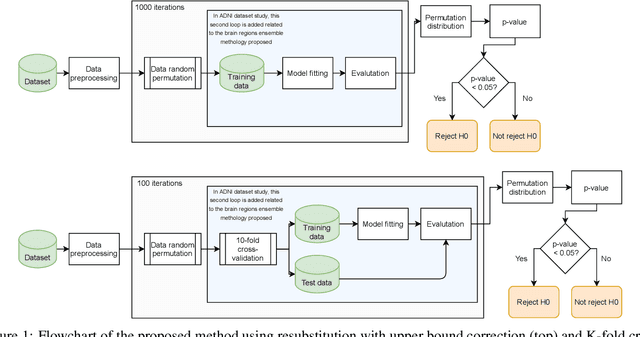
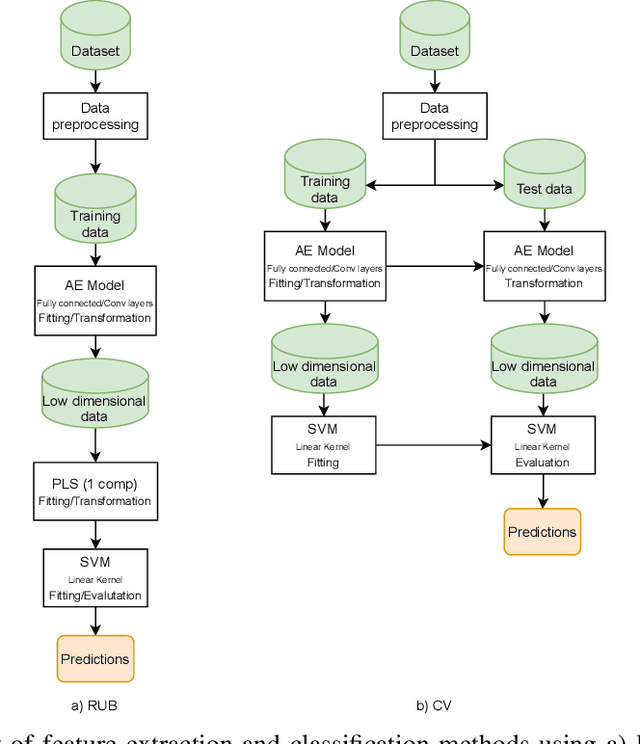
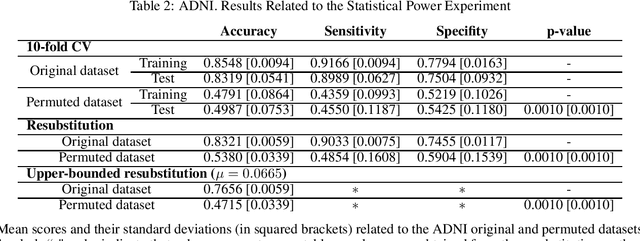
Abstract:Discriminative analysis in neuroimaging by means of deep/machine learning techniques is usually tested with validation techniques, whereas the associated statistical significance remains largely under-developed due to their computational complexity. In this work, a non-parametric framework is proposed that estimates the statistical significance of classifications using deep learning architectures. In particular, a combination of autoencoders (AE) and support vector machines (SVM) is applied to: (i) a one-condition, within-group designs often of normal controls (NC) and; (ii) a two-condition, between-group designs which contrast, for example, Alzheimer's disease (AD) patients with NC (the extension to multi-class analyses is also included). A random-effects inference based on a label permutation test is proposed in both studies using cross-validation (CV) and resubstitution with upper bound correction (RUB) as validation methods. This allows both false positives and classifier overfitting to be detected as well as estimating the statistical power of the test. Several experiments were carried out using the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset, the Dominantly Inherited Alzheimer Network (DIAN) dataset, and a MCI prediction dataset. We found in the permutation test that CV and RUB methods offer a false positive rate close to the significance level and an acceptable statistical power (although lower using cross-validation). A large separation between training and test accuracies using CV was observed, especially in one-condition designs. This implies a low generalization ability as the model fitted in training is not informative with respect to the test set. We propose as solution by applying RUB, whereby similar results are obtained to those of the CV test set, but considering the whole set and with a lower computational cost per iteration.
Ensemble Deep Learning on Large, Mixed-Site fMRI Datasets in Autism and Other Tasks
Feb 14, 2020



Abstract:Deep learning models for MRI classification face two recurring problems: they are typically limited by low sample size, and are abstracted by their own complexity (the "black box problem"). In this paper, we train a convolutional neural network (CNN) with the largest multi-source, functional MRI (fMRI) connectomic dataset ever compiled, consisting of 43,858 datapoints. We apply this model to a cross-sectional comparison of autism (ASD) vs typically developing (TD) controls that has proved difficult to characterise with inferential statistics. To contextualise these findings, we additionally perform classifications of gender and task vs rest. Employing class-balancing to build a training set, we trained 3$\times$300 modified CNNs in an ensemble model to classify fMRI connectivity matrices with overall AUROCs of 0.6774, 0.7680, and 0.9222 for ASD vs TD, gender, and task vs rest, respectively. Additionally, we aim to address the black box problem in this context using two visualization methods. First, class activation maps show which functional connections of the brain our models focus on when performing classification. Second, by analyzing maximal activations of the hidden layers, we were also able to explore how the model organizes a large and mixed-centre dataset, finding that it dedicates specific areas of its hidden layers to processing different covariates of data (depending on the independent variable analyzed), and other areas to mix data from different sources. Our study finds that deep learning models that distinguish ASD from TD controls focus broadly on temporal and cerebellar connections, with a particularly high focus on the right caudate nucleus and paracentral sulcus.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge