Deep Learning in current Neuroimaging: a multivariate approach with power and type I error control but arguable generalization ability
Paper and Code
Mar 30, 2021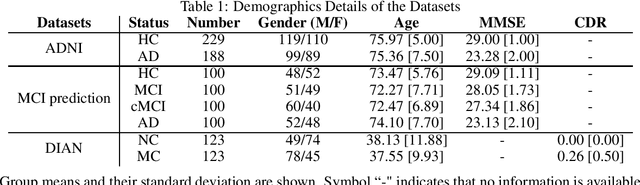
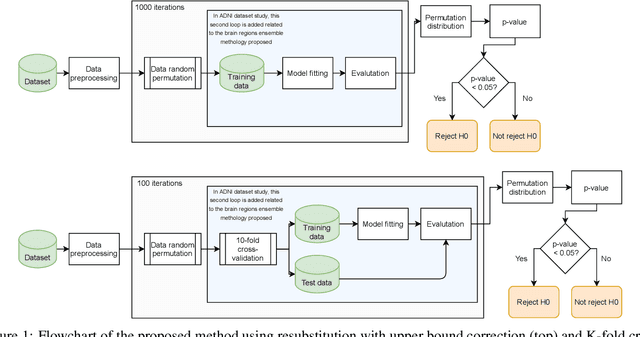
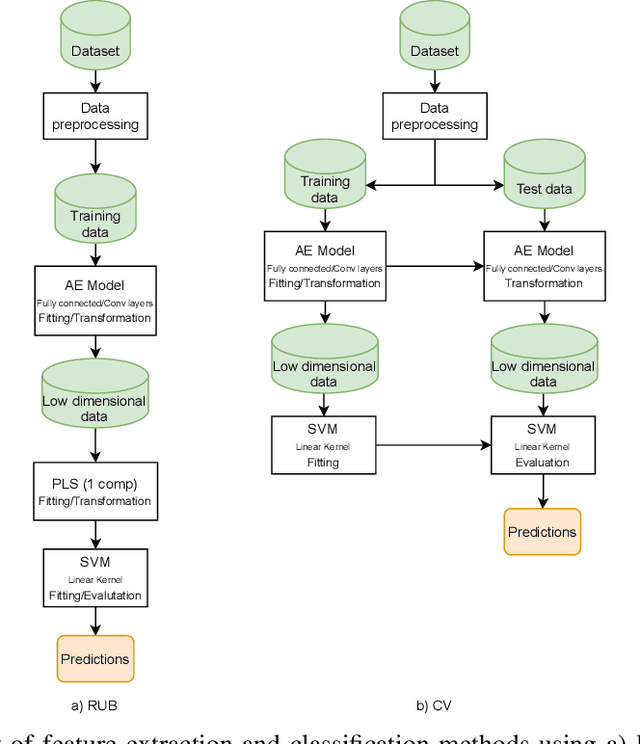
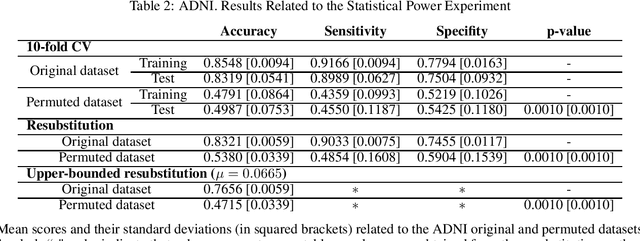
Discriminative analysis in neuroimaging by means of deep/machine learning techniques is usually tested with validation techniques, whereas the associated statistical significance remains largely under-developed due to their computational complexity. In this work, a non-parametric framework is proposed that estimates the statistical significance of classifications using deep learning architectures. In particular, a combination of autoencoders (AE) and support vector machines (SVM) is applied to: (i) a one-condition, within-group designs often of normal controls (NC) and; (ii) a two-condition, between-group designs which contrast, for example, Alzheimer's disease (AD) patients with NC (the extension to multi-class analyses is also included). A random-effects inference based on a label permutation test is proposed in both studies using cross-validation (CV) and resubstitution with upper bound correction (RUB) as validation methods. This allows both false positives and classifier overfitting to be detected as well as estimating the statistical power of the test. Several experiments were carried out using the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset, the Dominantly Inherited Alzheimer Network (DIAN) dataset, and a MCI prediction dataset. We found in the permutation test that CV and RUB methods offer a false positive rate close to the significance level and an acceptable statistical power (although lower using cross-validation). A large separation between training and test accuracies using CV was observed, especially in one-condition designs. This implies a low generalization ability as the model fitted in training is not informative with respect to the test set. We propose as solution by applying RUB, whereby similar results are obtained to those of the CV test set, but considering the whole set and with a lower computational cost per iteration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge