Matthew Leming
Multi-confound regression adversarial network for deep learning-based diagnosis on highly heterogenous clinical data
May 05, 2022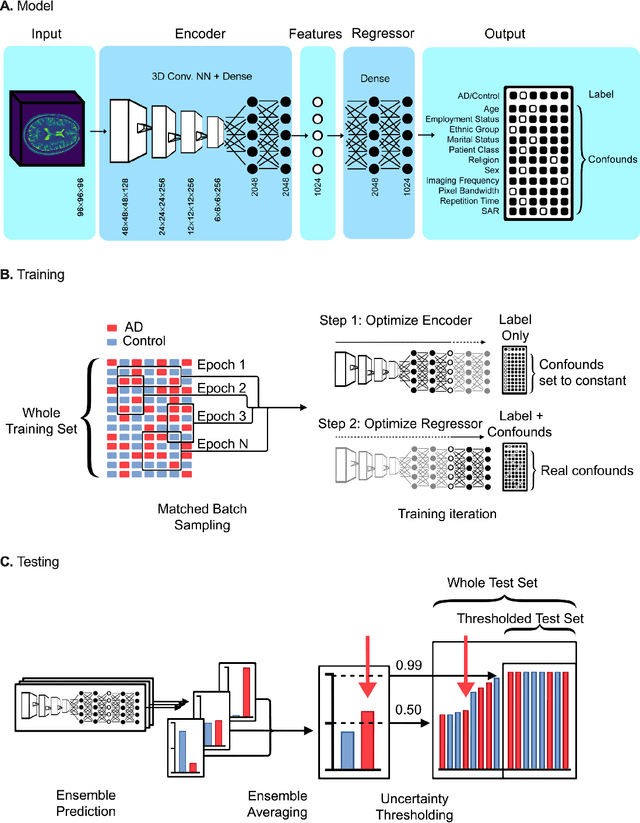
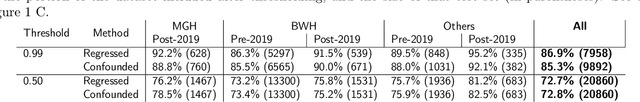
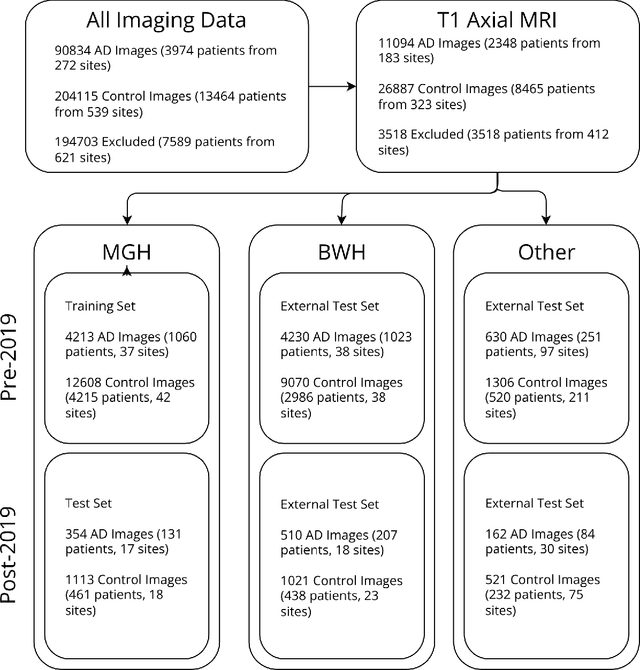
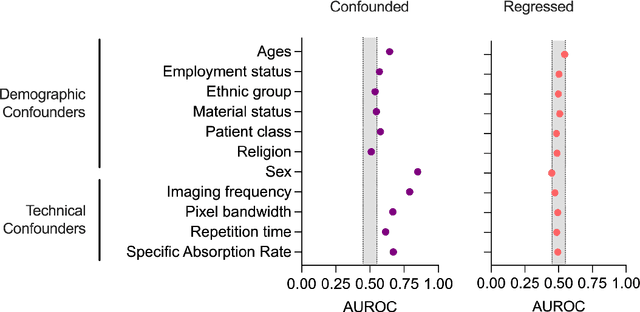
Abstract:Automated disease detection in medical images using deep learning holds promise to improve the diagnostic ability of radiologists, but routinely collected clinical data frequently contains technical and demographic confounding factors that differ between hospitals, negatively affecting the robustness of diagnostic deep learning models. Thus, there is a critical need for deep learning models that can train on imbalanced datasets without overfitting to site-specific confounding factors. In this work, we developed a novel deep learning architecture, MUCRAN (Multi-Confound Regression Adversarial Network), to train a deep learning model on highly heterogeneous clinical data while regressing demographic and technical confounding factors. We trained MUCRAN using 16,821 clinical T1 Axial brain MRIs collected from Massachusetts General Hospital before 2019 and tested it using post-2019 data to distinguish Alzheimer's disease (AD) patients, identified using both prescriptions of AD drugs and ICD codes, from a non-medicated control group. In external validation tests using MRI data from other hospitals, the model showed a robust performance of over 90% accuracy on newly collected data. This work shows the feasibility of deep learning-based diagnosis in real-world clinical data.
Single-participant structural connectivity matrices lead to greater accuracy in classification of participants than function in autism in MRI
May 27, 2020



Abstract:In this work, we introduce a technique of deriving symmetric connectivity matrices from regional histograms of grey-matter volume estimated from T1-weighted MRIs. We then validated the technique by inputting the connectivity matrices into a convolutional neural network (CNN) to classify between participants with autism and age-, motion-, and intracranial-volume-matched controls from six different databases (29,288 total connectomes, mean age = 30.72, range 0.42-78.00, including 1555 subjects with autism). We compared this method to similar classifications of the same participants using fMRI connectivity matrices as well as univariate estimates of grey-matter volumes. We further applied graph-theoretical metrics on output class activation maps to identify areas of the matrices that the CNN preferentially used to make the classification, focusing particularly on hubs. Our results gave AUROCs of 0.7298 (69.71% accuracy) when classifying by only structural connectivity, 0.6964 (67.72% accuracy) when classifying by only functional connectivity, and 0.7037 (66.43% accuracy) when classifying by univariate grey matter volumes. Combining structural and functional connectivities gave an AUROC of 0.7354 (69.40% accuracy). Graph analysis of class activation maps revealed no distinguishable network patterns for functional inputs, but did reveal localized differences between groups in bilateral Heschl's gyrus and upper vermis for structural connectivity. This work provides a simple means of feature extraction for inputting large numbers of structural MRIs into machine learning models.
Stochastic encoding of graphs in deep learning allows for complex analysis of gender classification in resting-state and task functional brain networks from the UK Biobank
Feb 25, 2020



Abstract:Classification of whole-brain functional connectivity MRI data with convolutional neural networks (CNNs) has shown promise, but the complexity of these models impedes understanding of which aspects of brain activity contribute to classification. While visualization techniques have been developed to interpret CNNs, bias inherent in the method of encoding abstract input data, as well as the natural variance of deep learning models, detract from the accuracy of these techniques. We introduce a stochastic encoding method in an ensemble of CNNs to classify functional connectomes by gender. We applied our method to resting-state and task data from the UK BioBank, using two visualization techniques to measure the salience of three brain networks involved in task- and resting-states, and their interaction. To regress confounding factors such as head motion, age, and intracranial volume, we introduced a multivariate balancing algorithm to ensure equal distributions of such covariates between classes in our data. We achieved a final AUROC of 0.8459. We found that resting-state data classifies more accurately than task data, with the inner salience network playing the most important role of the three networks overall in classification of resting-state data and connections to the central executive network in task data.
Ensemble Deep Learning on Large, Mixed-Site fMRI Datasets in Autism and Other Tasks
Feb 14, 2020



Abstract:Deep learning models for MRI classification face two recurring problems: they are typically limited by low sample size, and are abstracted by their own complexity (the "black box problem"). In this paper, we train a convolutional neural network (CNN) with the largest multi-source, functional MRI (fMRI) connectomic dataset ever compiled, consisting of 43,858 datapoints. We apply this model to a cross-sectional comparison of autism (ASD) vs typically developing (TD) controls that has proved difficult to characterise with inferential statistics. To contextualise these findings, we additionally perform classifications of gender and task vs rest. Employing class-balancing to build a training set, we trained 3$\times$300 modified CNNs in an ensemble model to classify fMRI connectivity matrices with overall AUROCs of 0.6774, 0.7680, and 0.9222 for ASD vs TD, gender, and task vs rest, respectively. Additionally, we aim to address the black box problem in this context using two visualization methods. First, class activation maps show which functional connections of the brain our models focus on when performing classification. Second, by analyzing maximal activations of the hidden layers, we were also able to explore how the model organizes a large and mixed-centre dataset, finding that it dedicates specific areas of its hidden layers to processing different covariates of data (depending on the independent variable analyzed), and other areas to mix data from different sources. Our study finds that deep learning models that distinguish ASD from TD controls focus broadly on temporal and cerebellar connections, with a particularly high focus on the right caudate nucleus and paracentral sulcus.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge