Juan M. Górriz
An explainable framework for the relationship between dementia and glucose metabolism patterns
Jan 28, 2026Abstract:High-dimensional neuroimaging data presents challenges for assessing neurodegenerative diseases due to complex non-linear relationships. Variational Autoencoders (VAEs) can encode scans into lower-dimensional latent spaces capturing disease-relevant features. We propose a semi-supervised VAE framework with a flexible similarity regularization term that aligns selected latent variables with clinical or biomarker measures of dementia progression. This allows adapting the similarity metric and supervised variables to specific goals or available data. We demonstrate the approach using PET scans from the Alzheimer's Disease Neuroimaging Initiative (ADNI), guiding the first latent dimension to align with a cognitive score. Using this supervised latent variable, we generate average reconstructions across levels of cognitive impairment. Voxel-wise GLM analysis reveals reduced metabolism in key regions, mainly the hippocampus, and within major Resting State Networks, particularly the Default Mode and Central Executive Networks. The remaining latent variables encode affine transformations and intensity variations, capturing confounds such as inter-subject variability and site effects. Our framework effectively extracts disease-related patterns aligned with established Alzheimer's biomarkers, offering an interpretable and adaptable tool for studying neurodegenerative progression.
A Review of Latent Representation Models in Neuroimaging
Dec 24, 2024



Abstract:Neuroimaging data, particularly from techniques like MRI or PET, offer rich but complex information about brain structure and activity. To manage this complexity, latent representation models - such as Autoencoders, Generative Adversarial Networks (GANs), and Latent Diffusion Models (LDMs) - are increasingly applied. These models are designed to reduce high-dimensional neuroimaging data to lower-dimensional latent spaces, where key patterns and variations related to brain function can be identified. By modeling these latent spaces, researchers hope to gain insights into the biology and function of the brain, including how its structure changes with age or disease, or how it encodes sensory information, predicts and adapts to new inputs. This review discusses how these models are used for clinical applications, like disease diagnosis and progression monitoring, but also for exploring fundamental brain mechanisms such as active inference and predictive coding. These approaches provide a powerful tool for both understanding and simulating the brain's complex computational tasks, potentially advancing our knowledge of cognition, perception, and neural disorders.
Convolutional Neural Networks for Neuroimaging in Parkinson's Disease: Is Preprocessing Needed?
Nov 21, 2023



Abstract:Spatial and intensity normalization are nowadays a prerequisite for neuroimaging analysis. Influenced by voxel-wise and other univariate comparisons, where these corrections are key, they are commonly applied to any type of analysis and imaging modalities. Nuclear imaging modalities such as PET-FDG or FP-CIT SPECT, a common modality used in Parkinson's Disease diagnosis, are especially dependent on intensity normalization. However, these steps are computationally expensive and furthermore, they may introduce deformations in the images, altering the information contained in them. Convolutional Neural Networks (CNNs), for their part, introduce position invariance to pattern recognition, and have been proven to classify objects regardless of their orientation, size, angle, etc. Therefore, a question arises: how well can CNNs account for spatial and intensity differences when analysing nuclear brain imaging? Are spatial and intensity normalization still needed? To answer this question, we have trained four different CNN models based on well-established architectures, using or not different spatial and intensity normalization preprocessing. The results show that a sufficiently complex model such as our three-dimensional version of the ALEXNET can effectively account for spatial differences, achieving a diagnosis accuracy of 94.1% with an area under the ROC curve of 0.984. The visualization of the differences via saliency maps shows that these models are correctly finding patterns that match those found in the literature, without the need of applying any complex spatial normalization procedure. However, the intensity normalization -- and its type -- is revealed as very influential in the results and accuracy of the trained model, and therefore must be well accounted.
* 19 pages, 7 figures
Probabilistic combination of eigenlungs-based classifiers for COVID-19 diagnosis in chest CT images
Mar 04, 2021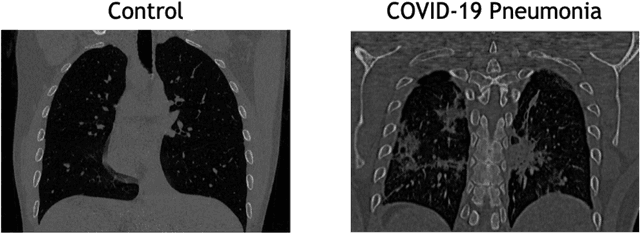
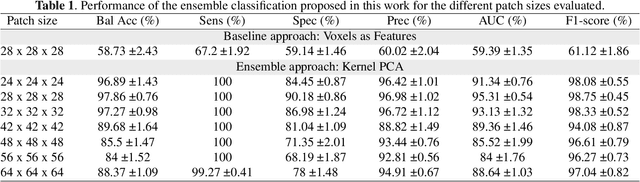
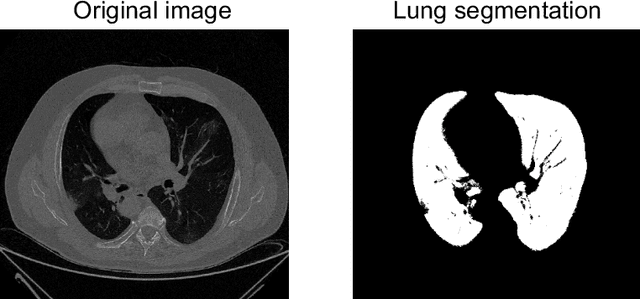
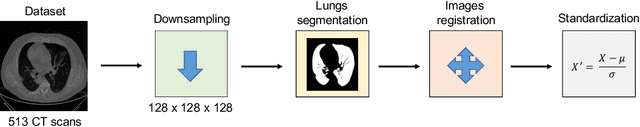
Abstract:The outbreak of the COVID-19 (Coronavirus disease 2019) pandemic has changed the world. According to the World Health Organization (WHO), there have been more than 100 million confirmed cases of COVID-19, including more than 2.4 million deaths. It is extremely important the early detection of the disease, and the use of medical imaging such as chest X-ray (CXR) and chest Computed Tomography (CCT) have proved to be an excellent solution. However, this process requires clinicians to do it within a manual and time-consuming task, which is not ideal when trying to speed up the diagnosis. In this work, we propose an ensemble classifier based on probabilistic Support Vector Machine (SVM) in order to identify pneumonia patterns while providing information about the reliability of the classification. Specifically, each CCT scan is divided into cubic patches and features contained in each one of them are extracted by applying kernel PCA. The use of base classifiers within an ensemble allows our system to identify the pneumonia patterns regardless of their size or location. Decisions of each individual patch are then combined into a global one according to the reliability of each individual classification: the lower the uncertainty, the higher the contribution. Performance is evaluated in a real scenario, yielding an accuracy of 97.86%. The large performance obtained and the simplicity of the system (use of deep learning in CCT images would result in a huge computational cost) evidence the applicability of our proposal in a real-world environment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge