Sai Ho Ling
SA$^{2}$Net: Scale-Adaptive Structure-Affinity Transformation for Spine Segmentation from Ultrasound Volume Projection Imaging
Oct 30, 2025Abstract:Spine segmentation, based on ultrasound volume projection imaging (VPI), plays a vital role for intelligent scoliosis diagnosis in clinical applications. However, this task faces several significant challenges. Firstly, the global contextual knowledge of spines may not be well-learned if we neglect the high spatial correlation of different bone features. Secondly, the spine bones contain rich structural knowledge regarding their shapes and positions, which deserves to be encoded into the segmentation process. To address these challenges, we propose a novel scale-adaptive structure-aware network (SA$^{2}$Net) for effective spine segmentation. First, we propose a scale-adaptive complementary strategy to learn the cross-dimensional long-distance correlation features for spinal images. Second, motivated by the consistency between multi-head self-attention in Transformers and semantic level affinity, we propose structure-affinity transformation to transform semantic features with class-specific affinity and combine it with a Transformer decoder for structure-aware reasoning. In addition, we adopt a feature mixing loss aggregation method to enhance model training. This method improves the robustness and accuracy of the segmentation process. The experimental results demonstrate that our SA$^{2}$Net achieves superior segmentation performance compared to other state-of-the-art methods. Moreover, the adaptability of SA$^{2}$Net to various backbones enhances its potential as a promising tool for advanced scoliosis diagnosis using intelligent spinal image analysis. The code and experimental demo are available at https://github.com/taetiseo09/SA2Net.
Multimodal Learning for Non-small Cell Lung Cancer Prognosis
Nov 07, 2022Abstract:This paper focuses on the task of survival time analysis for lung cancer. Although much progress has been made in this problem in recent years, the performance of existing methods is still far from satisfactory. Traditional and some deep learning-based survival time analyses for lung cancer are mostly based on textual clinical information such as staging, age, histology, etc. Unlike existing methods that predicting on the single modality, we observe that a human clinician usually takes multimodal data such as text clinical data and visual scans to estimate survival time. Motivated by this, in this work, we contribute a smart cross-modality network for survival analysis network named Lite-ProSENet that simulates a human's manner of decision making. Extensive experiments were conducted using data from 422 NSCLC patients from The Cancer Imaging Archive (TCIA). The results show that our Lite-ProSENet outperforms favorably again all comparison methods and achieves the new state of the art with the 89.3% on concordance. The code will be made publicly available.
Automatic Diagnosis of Myocarditis Disease in Cardiac MRI Modality using Deep Transformers and Explainable Artificial Intelligence
Oct 26, 2022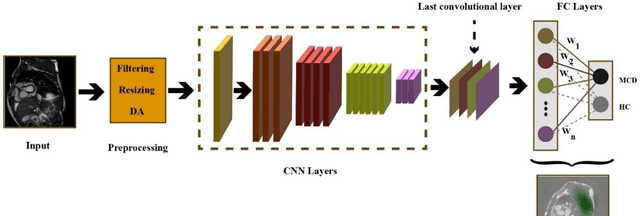
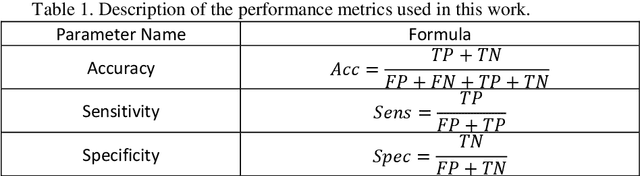
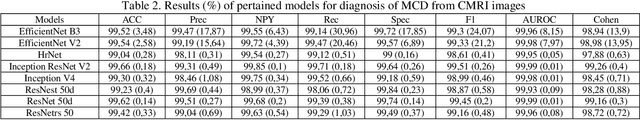
Abstract:Myocarditis is among the most important cardiovascular diseases (CVDs), endangering the health of many individuals by damaging the myocardium. Microbes and viruses, such as HIV, play a vital role in myocarditis disease (MCD) incidence. Lack of MCD diagnosis in the early stages is associated with irreversible complications. Cardiac magnetic resonance imaging (CMRI) is highly popular among cardiologists to diagnose CVDs. In this paper, a deep learning (DL) based computer-aided diagnosis system (CADS) is presented for the diagnosis of MCD using CMRI images. The proposed CADS includes dataset, preprocessing, feature extraction, classification, and post-processing steps. First, the Z-Alizadeh dataset was selected for the experiments. The preprocessing step included noise removal, image resizing, and data augmentation (DA). In this step, CutMix, and MixUp techniques were used for the DA. Then, the most recent pre-trained and transformers models were used for feature extraction and classification using CMRI images. Our results show high performance for the detection of MCD using transformer models compared with the pre-trained architectures. Among the DL architectures, Turbulence Neural Transformer (TNT) architecture achieved an accuracy of 99.73% with 10-fold cross-validation strategy. Explainable-based Grad Cam method is used to visualize the MCD suspected areas in CMRI images.
Automated Diagnosis of Cardiovascular Diseases from Cardiac Magnetic Resonance Imaging Using Deep Learning Models: A Review
Oct 26, 2022Abstract:In recent years, cardiovascular diseases (CVDs) have become one of the leading causes of mortality globally. CVDs appear with minor symptoms and progressively get worse. The majority of people experience symptoms such as exhaustion, shortness of breath, ankle swelling, fluid retention, and other symptoms when starting CVD. Coronary artery disease (CAD), arrhythmia, cardiomyopathy, congenital heart defect (CHD), mitral regurgitation, and angina are the most common CVDs. Clinical methods such as blood tests, electrocardiography (ECG) signals, and medical imaging are the most effective methods used for the detection of CVDs. Among the diagnostic methods, cardiac magnetic resonance imaging (CMR) is increasingly used to diagnose, monitor the disease, plan treatment and predict CVDs. Coupled with all the advantages of CMR data, CVDs diagnosis is challenging for physicians due to many slices of data, low contrast, etc. To address these issues, deep learning (DL) techniques have been employed to the diagnosis of CVDs using CMR data, and much research is currently being conducted in this field. This review provides an overview of the studies performed in CVDs detection using CMR images and DL techniques. The introduction section examined CVDs types, diagnostic methods, and the most important medical imaging techniques. In the following, investigations to detect CVDs using CMR images and the most significant DL methods are presented. Another section discussed the challenges in diagnosing CVDs from CMR data. Next, the discussion section discusses the results of this review, and future work in CVDs diagnosis from CMR images and DL techniques are outlined. The most important findings of this study are presented in the conclusion section.
Automatic Autism Spectrum Disorder Detection Using Artificial Intelligence Methods with MRI Neuroimaging: A Review
Jun 20, 2022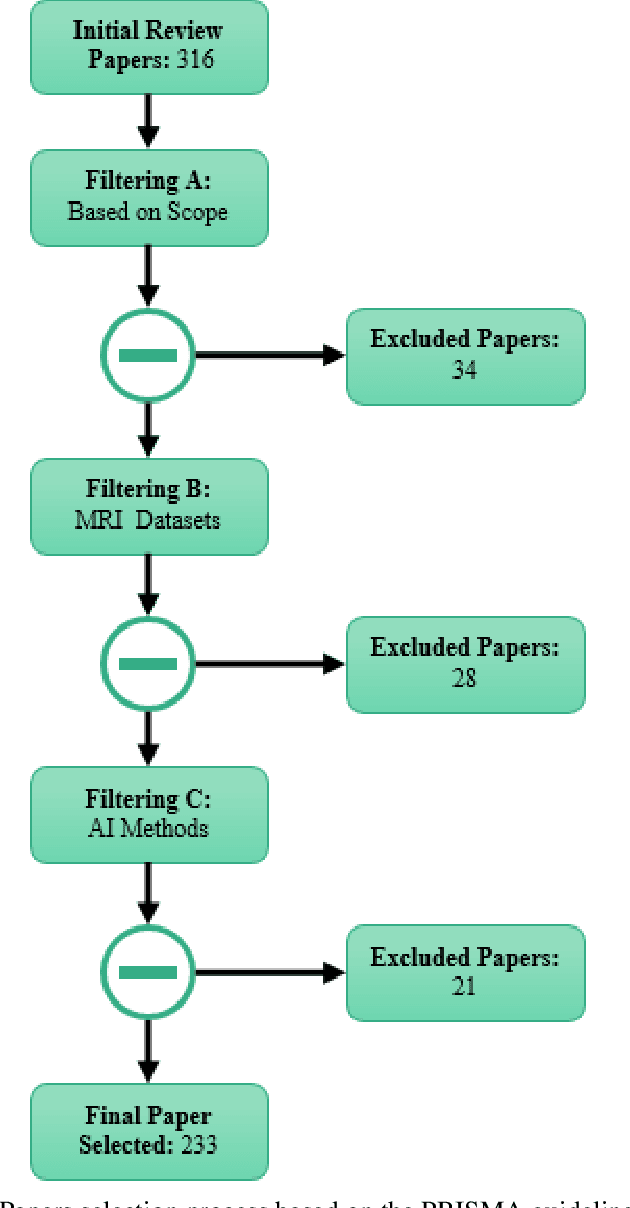

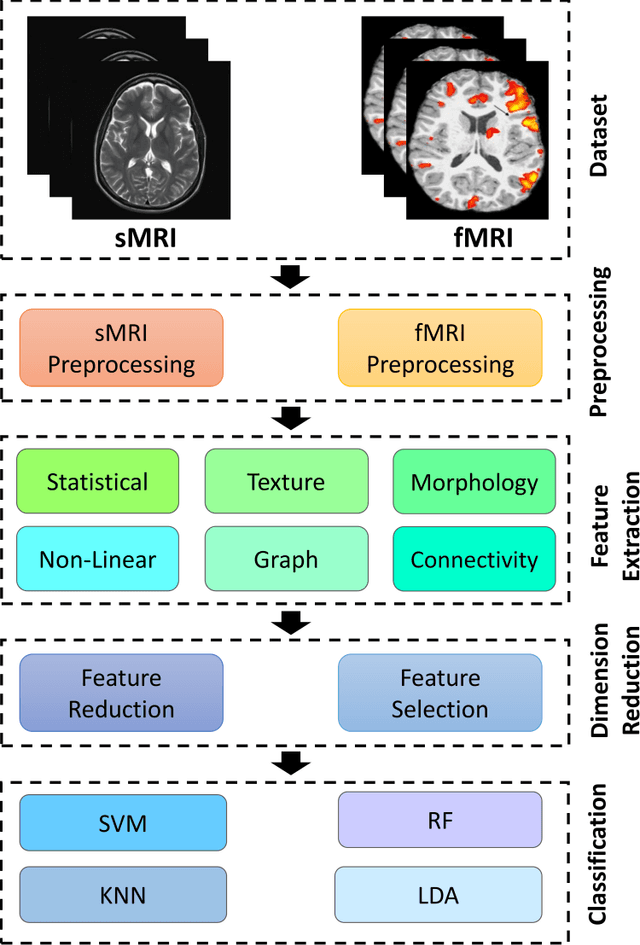
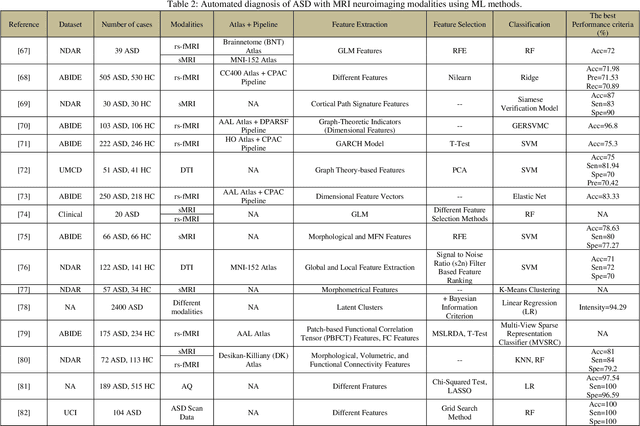
Abstract:Autism spectrum disorder (ASD) is a brain condition characterized by diverse signs and symptoms that appear in early childhood. ASD is also associated with communication deficits and repetitive behavior in affected individuals. Various ASD detection methods have been developed, including neuroimaging modalities and psychological tests. Among these methods, magnetic resonance imaging (MRI) imaging modalities are of paramount importance to physicians. Clinicians rely on MRI modalities to diagnose ASD accurately. The MRI modalities are non-invasive methods that include functional (fMRI) and structural (sMRI) neuroimaging methods. However, the process of diagnosing ASD with fMRI and sMRI for specialists is often laborious and time-consuming; therefore, several computer-aided design systems (CADS) based on artificial intelligence (AI) have been developed to assist the specialist physicians. Conventional machine learning (ML) and deep learning (DL) are the most popular schemes of AI used for diagnosing ASD. This study aims to review the automated detection of ASD using AI. We review several CADS that have been developed using ML techniques for the automated diagnosis of ASD using MRI modalities. There has been very limited work on the use of DL techniques to develop automated diagnostic models for ASD. A summary of the studies developed using DL is provided in the appendix. Then, the challenges encountered during the automated diagnosis of ASD using MRI and AI techniques are described in detail. Additionally, a graphical comparison of studies using ML and DL to diagnose ASD automatically is discussed. We conclude by suggesting future approaches to detecting ASDs using AI techniques and MRI neuroimaging.
DeepMMSA: A Novel Multimodal Deep Learning Method for Non-small Cell Lung Cancer Survival Analysis
Jun 12, 2021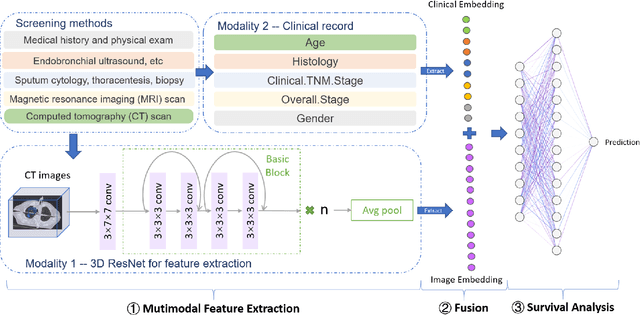
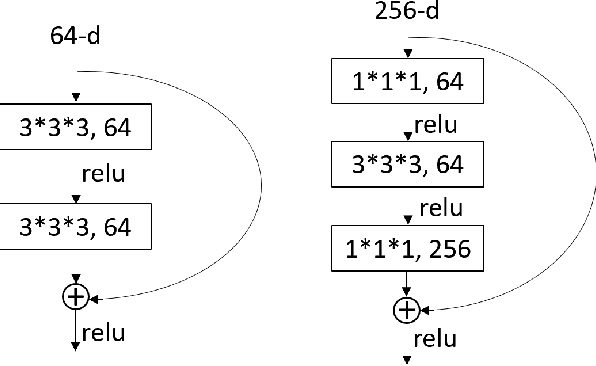
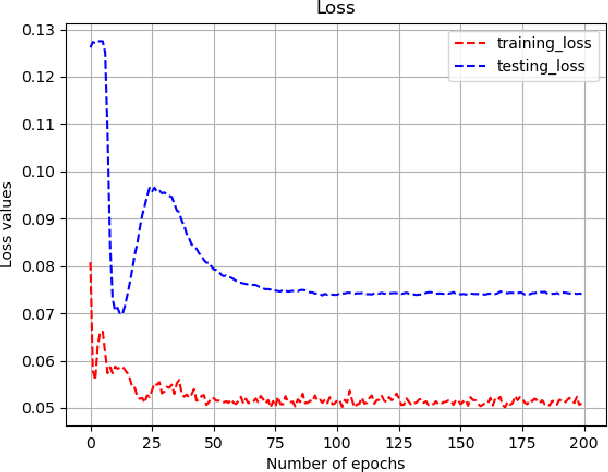
Abstract:Lung cancer is the leading cause of cancer death worldwide. The critical reason for the deaths is delayed diagnosis and poor prognosis. With the accelerated development of deep learning techniques, it has been successfully applied extensively in many real-world applications, including health sectors such as medical image interpretation and disease diagnosis. By combining more modalities that being engaged in the processing of information, multimodal learning can extract better features and improve predictive ability. The conventional methods for lung cancer survival analysis normally utilize clinical data and only provide a statistical probability. To improve the survival prediction accuracy and help prognostic decision-making in clinical practice for medical experts, we for the first time propose a multimodal deep learning method for non-small cell lung cancer (NSCLC) survival analysis, named DeepMMSA. This method leverages CT images in combination with clinical data, enabling the abundant information hold within medical images to be associate with lung cancer survival information. We validate our method on the data of 422 NSCLC patients from The Cancer Imaging Archive (TCIA). Experimental results support our hypothesis that there is an underlying relationship between prognostic information and radiomic images. Besides, quantitative results showing that the established multimodal model can be applied to traditional method and has the potential to break bottleneck of existing methods and increase the the percentage of concordant pairs(right predicted pairs) in overall population by 4%.
Bone Feature Segmentation in Ultrasound Spine Image with Robustness to Speckle and Regular Occlusion Noise
Oct 08, 2020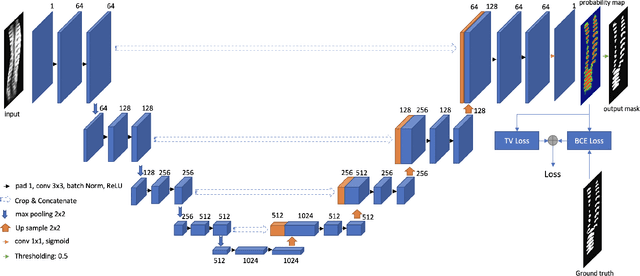
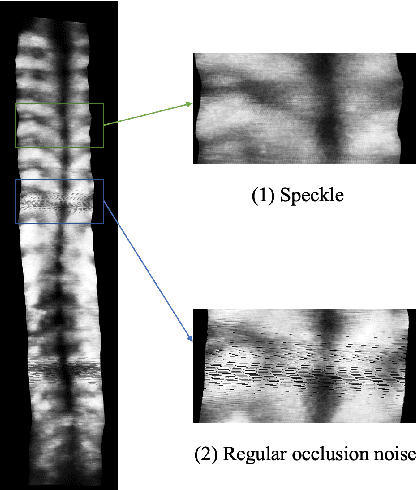
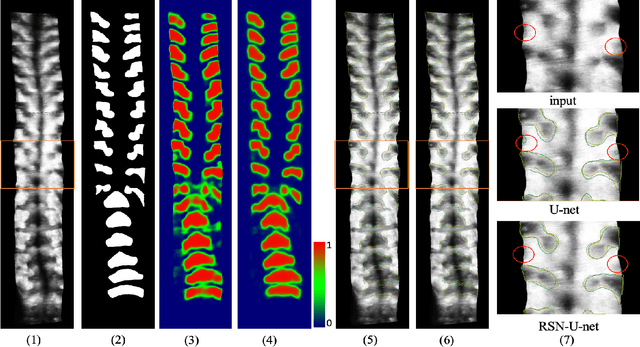
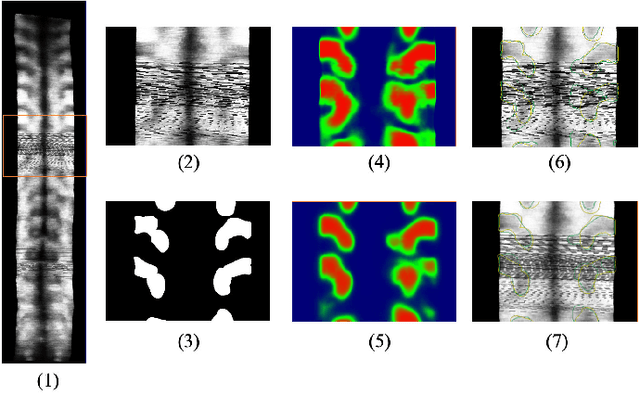
Abstract:3D ultrasound imaging shows great promise for scoliosis diagnosis thanks to its low-costing, radiation-free and real-time characteristics. The key to accessing scoliosis by ultrasound imaging is to accurately segment the bone area and measure the scoliosis degree based on the symmetry of the bone features. The ultrasound images tend to contain many speckles and regular occlusion noise which is difficult, tedious and time-consuming for experts to find out the bony feature. In this paper, we propose a robust bone feature segmentation method based on the U-net structure for ultrasound spine Volume Projection Imaging (VPI) images. The proposed segmentation method introduces a total variance loss to reduce the sensitivity of the model to small-scale and regular occlusion noise. The proposed approach improves 2.3% of Dice score and 1% of AUC score as compared with the u-net model and shows high robustness to speckle and regular occlusion noise.
Multi-level CNN for lung nodule classification with Gaussian Process assisted hyperparameter optimization
Jan 02, 2019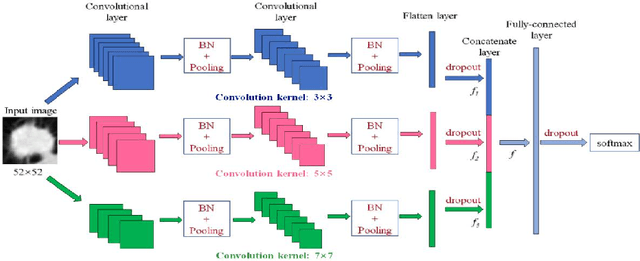


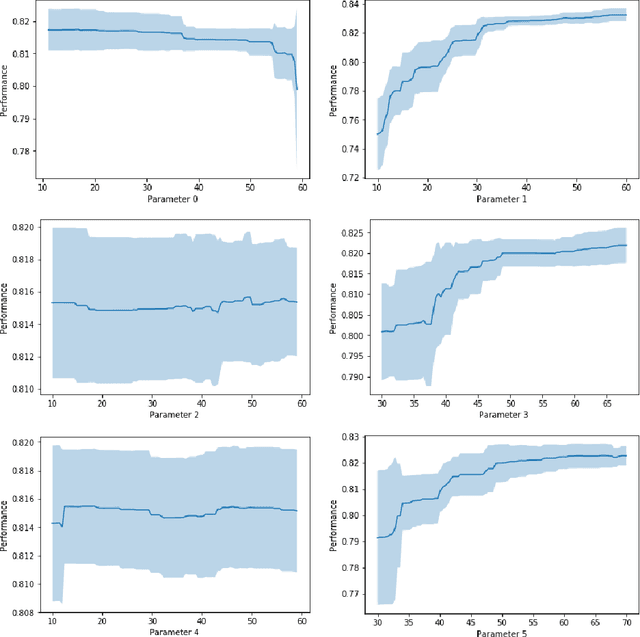
Abstract:This paper investigates lung nodule classification by using deep neural networks (DNNs). Hyperparameter optimization in DNNs is a computationally expensive problem, where evaluating a hyperparameter configuration may take several hours or even days. Bayesian optimization has been recently introduced for the automatically searching of optimal hyperparameter configurations of DNNs. It applies probabilistic surrogate models to approximate the validation error function of hyperparameter configurations, such as Gaussian processes, and reduce the computational complexity to a large extent. However, most existing surrogate models adopt stationary covariance functions to measure the difference between hyperparameter points based on spatial distance without considering its spatial locations. This distance-based assumption together with the condition of constant smoothness throughout the whole hyperparameter search space clearly violates the property that the points far away from optimal points usually get similarly poor performance even though each two of them have huge spatial distance between them. In this paper, a non-stationary kernel is proposed which allows the surrogate model to adapt to functions whose smoothness varies with the spatial location of inputs, and a multi-level convolutional neural network (ML-CNN) is built for lung nodule classification whose hyperparameter configuration is optimized by using the proposed non-stationary kernel based Gaussian surrogate model. Our algorithm searches the surrogate for optimal setting via hyperparameter importance based evolutionary strategy, and the experiments demonstrate our algorithm outperforms manual tuning and well-established hyperparameter optimization methods such as Random search, Gaussian processes with stationary kernels, and recently proposed Hyperparameter Optimization via RBF and Dynamic coordinate search.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge