F. J. Martinez-Murcia
Statistical Agnostic Regression: a machine learning method to validate regression models
Feb 23, 2024



Abstract:Regression analysis is a central topic in statistical modeling, aiming to estimate the relationships between a dependent variable, commonly referred to as the response variable, and one or more independent variables, i.e., explanatory variables. Linear regression is by far the most popular method for performing this task in several fields of research, such as prediction, forecasting, or causal inference. Beyond various classical methods to solve linear regression problems, such as Ordinary Least Squares, Ridge, or Lasso regressions - which are often the foundation for more advanced machine learning (ML) techniques - the latter have been successfully applied in this scenario without a formal definition of statistical significance. At most, permutation or classical analyses based on empirical measures (e.g., residuals or accuracy) have been conducted to reflect the greater ability of ML estimations for detection. In this paper, we introduce a method, named Statistical Agnostic Regression (SAR), for evaluating the statistical significance of an ML-based linear regression based on concentration inequalities of the actual risk using the analysis of the worst case. To achieve this goal, similar to the classification problem, we define a threshold to establish that there is sufficient evidence with a probability of at least 1-eta to conclude that there is a linear relationship in the population between the explanatory (feature) and the response (label) variables. Simulations in only two dimensions demonstrate the ability of the proposed agnostic test to provide a similar analysis of variance given by the classical $F$ test for the slope parameter.
Revealing Patterns of Symptomatology in Parkinson's Disease: A Latent Space Analysis with 3D Convolutional Autoencoders
May 11, 2023Abstract:This work proposes the use of 3D convolutional variational autoencoders (CVAEs) to trace the changes and symptomatology produced by neurodegeneration in Parkinson's disease (PD). In this work, we present a novel approach to detect and quantify changes in dopamine transporter (DaT) concentration and its spatial patterns using 3D CVAEs on Ioflupane (FPCIT) imaging. Our approach leverages the power of deep learning to learn a low-dimensional representation of the brain imaging data, which then is linked to different symptom categories using regression algorithms. We demonstrate the effectiveness of our approach on a dataset of PD patients and healthy controls, and show that general symptomatology (UPDRS) is linked to a d-dimensional decomposition via the CVAE with R2>0.25. Our work shows the potential of representation learning not only in early diagnosis but in understanding neurodegeneration processes and symptomatology.
Uncertainty-driven ensembles of deep architectures for multiclass classification. Application to COVID-19 diagnosis in chest X-ray images
Nov 27, 2020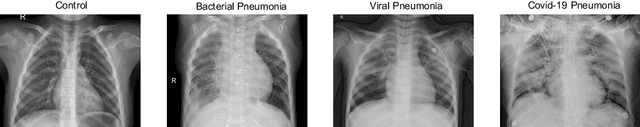
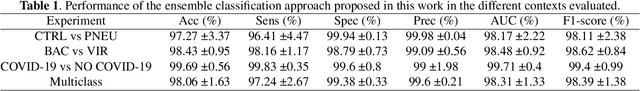
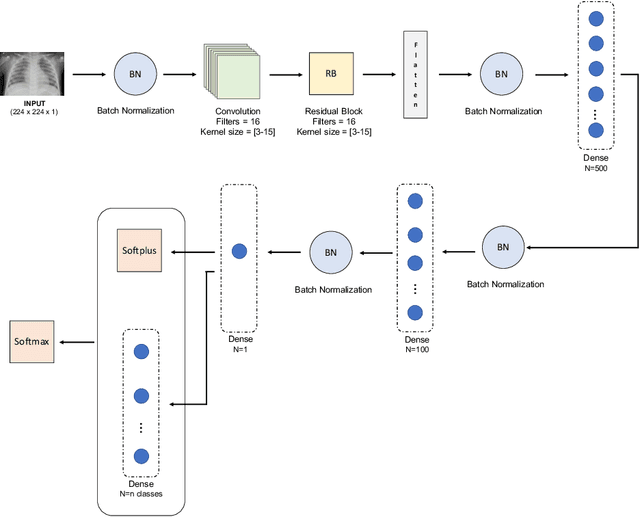
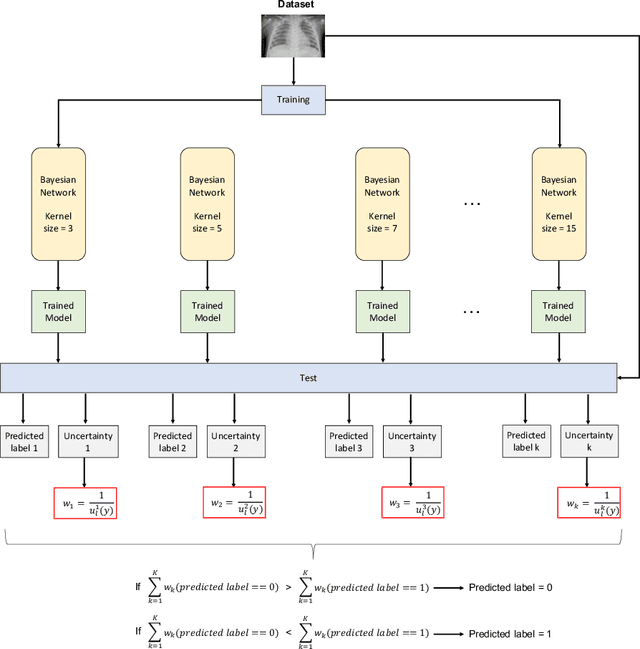
Abstract:Respiratory diseases kill million of people each year. Diagnosis of these pathologies is a manual, time-consuming process that has inter and intra-observer variability, delaying diagnosis and treatment. The recent COVID-19 pandemic has demonstrated the need of developing systems to automatize the diagnosis of pneumonia, whilst Convolutional Neural Network (CNNs) have proved to be an excellent option for the automatic classification of medical images. However, given the need of providing a confidence classification in this context it is crucial to quantify the reliability of the model's predictions. In this work, we propose a multi-level ensemble classification system based on a Bayesian Deep Learning approach in order to maximize performance while quantifying the uncertainty of each classification decision. This tool combines the information extracted from different architectures by weighting their results according to the uncertainty of their predictions. Performance of the Bayesian network is evaluated in a real scenario where simultaneously differentiating between four different pathologies: control vs bacterial pneumonia vs viral pneumonia vs COVID-19 pneumonia. A three-level decision tree is employed to divide the 4-class classification into three binary classifications, yielding an accuracy of 98.06% and overcoming the results obtained by recent literature. The reduced preprocessing needed for obtaining this high performance, in addition to the information provided about the reliability of the predictions evidence the applicability of the system to be used as an aid for clinicians.
A Neural Approach to Ordinal Regression for the Preventive Assessment of Developmental Dyslexia
Feb 06, 2020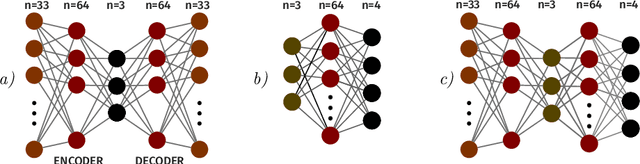
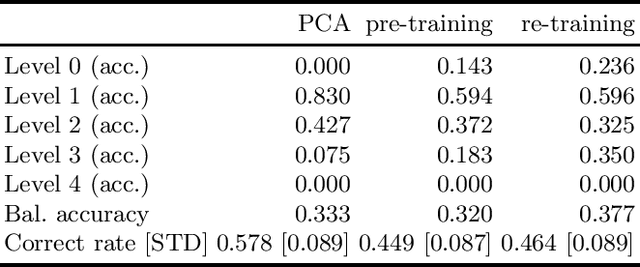
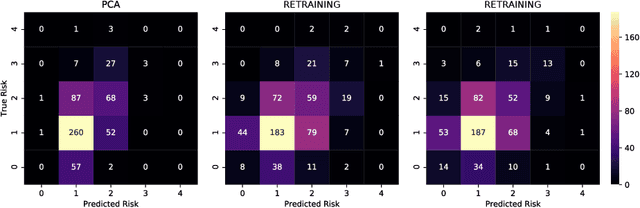
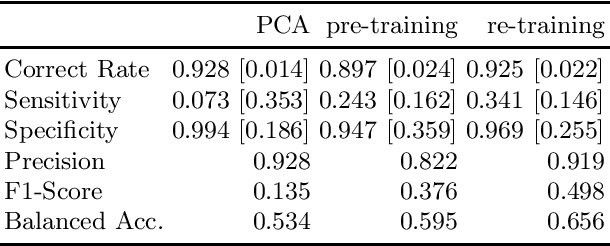
Abstract:Developmental Dyslexia (DD) is a learning disability related to the acquisition of reading skills that affects about 5% of the population. DD can have an enormous impact on the intellectual and personal development of affected children, so early detection is key to implementing preventive strategies for teaching language. Research has shown that there may be biological underpinnings to DD that affect phoneme processing, and hence these symptoms may be identifiable before reading ability is acquired, allowing for early intervention. In this paper we propose a new methodology to assess the risk of DD before students learn to read. For this purpose, we propose a mixed neural model that calculates risk levels of dyslexia from tests that can be completed at the age of 5 years. Our method first trains an auto-encoder, and then combines the trained encoder with an optimized ordinal regression neural network devised to ensure consistency of predictions. Our experiments show that the system is able to detect unaffected subjects two years before it can assess the risk of DD based mainly on phonological processing, giving a specificity of 0.969 and a correct rate of more than 0.92. In addition, the trained encoder can be used to transform test results into an interpretable subject spatial distribution that facilitates risk assessment and validates methodology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge