Tianqi Xiang
EFSI-DETR: Efficient Frequency-Semantic Integration for Real-Time Small Object Detection in UAV Imagery
Jan 26, 2026Abstract:Real-time small object detection in Unmanned Aerial Vehicle (UAV) imagery remains challenging due to limited feature representation and ineffective multi-scale fusion. Existing methods underutilize frequency information and rely on static convolutional operations, which constrain the capacity to obtain rich feature representations and hinder the effective exploitation of deep semantic features. To address these issues, we propose EFSI-DETR, a novel detection framework that integrates efficient semantic feature enhancement with dynamic frequency-spatial guidance. EFSI-DETR comprises two main components: (1) a Dynamic Frequency-Spatial Unified Synergy Network (DyFusNet) that jointly exploits frequency and spatial cues for robust multi-scale feature fusion, (2) an Efficient Semantic Feature Concentrator (ESFC) that enables deep semantic extraction with minimal computational cost. Furthermore, a Fine-grained Feature Retention (FFR) strategy is adopted to incorporate spatially rich shallow features during fusion to preserve fine-grained details, crucial for small object detection in UAV imagery. Extensive experiments on VisDrone and CODrone benchmarks demonstrate that our EFSI-DETR achieves the state-of-the-art performance with real-time efficiency, yielding improvement of \textbf{1.6}\% and \textbf{5.8}\% in AP and AP$_{s}$ on VisDrone, while obtaining \textbf{188} FPS inference speed on a single RTX 4090 GPU.
MOC: Meta-Optimized Classifier for Few-Shot Whole Slide Image Classification
Aug 13, 2025Abstract:Recent advances in histopathology vision-language foundation models (VLFMs) have shown promise in addressing data scarcity for whole slide image (WSI) classification via zero-shot adaptation. However, these methods remain outperformed by conventional multiple instance learning (MIL) approaches trained on large datasets, motivating recent efforts to enhance VLFM-based WSI classification through fewshot learning paradigms. While existing few-shot methods improve diagnostic accuracy with limited annotations, their reliance on conventional classifier designs introduces critical vulnerabilities to data scarcity. To address this problem, we propose a Meta-Optimized Classifier (MOC) comprising two core components: (1) a meta-learner that automatically optimizes a classifier configuration from a mixture of candidate classifiers and (2) a classifier bank housing diverse candidate classifiers to enable a holistic pathological interpretation. Extensive experiments demonstrate that MOC outperforms prior arts in multiple few-shot benchmarks. Notably, on the TCGA-NSCLC benchmark, MOC improves AUC by 10.4% over the state-of-the-art few-shot VLFM-based methods, with gains up to 26.25% under 1-shot conditions, offering a critical advancement for clinical deployments where diagnostic training data is severely limited. Code is available at https://github.com/xmed-lab/MOC.
Adaptive Prototype Model for Attribute-based Multi-label Few-shot Action Recognition
Feb 18, 2025Abstract:In real-world action recognition systems, incorporating more attributes helps achieve a more comprehensive understanding of human behavior. However, using a single model to simultaneously recognize multiple attributes can lead to a decrease in accuracy. In this work, we propose a novel method i.e. Adaptive Attribute Prototype Model (AAPM) for human action recognition, which captures rich action-relevant attribute information and strikes a balance between accuracy and robustness. Firstly, we introduce the Text-Constrain Module (TCM) to incorporate textual information from potential labels, and constrain the construction of different attributes prototype representations. In addition, we explore the Attribute Assignment Method (AAM) to address the issue of training bias and increase robustness during the training process.Furthermore, we construct a new video dataset with attribute-based multi-label called Multi-Kinetics for evaluation, which contains various attribute labels (e.g. action, scene, object, etc.) related to human behavior. Extensive experiments demonstrate that our AAPM achieves the state-of-the-art performance in both attribute-based multi-label few-shot action recognition and single-label few-shot action recognition. The project and dataset are available at an anonymous account https://github.com/theAAPM/AAPM
DiffCMR: Fast Cardiac MRI Reconstruction with Diffusion Probabilistic Models
Dec 08, 2023



Abstract:Performing magnetic resonance imaging (MRI) reconstruction from under-sampled k-space data can accelerate the procedure to acquire MRI scans and reduce patients' discomfort. The reconstruction problem is usually formulated as a denoising task that removes the noise in under-sampled MRI image slices. Although previous GAN-based methods have achieved good performance in image denoising, they are difficult to train and require careful tuning of hyperparameters. In this paper, we propose a novel MRI denoising framework DiffCMR by leveraging conditional denoising diffusion probabilistic models. Specifically, DiffCMR perceives conditioning signals from the under-sampled MRI image slice and generates its corresponding fully-sampled MRI image slice. During inference, we adopt a multi-round ensembling strategy to stabilize the performance. We validate DiffCMR with cine reconstruction and T1/T2 mapping tasks on MICCAI 2023 Cardiac MRI Reconstruction Challenge (CMRxRecon) dataset. Results show that our method achieves state-of-the-art performance, exceeding previous methods by a significant margin. Code is available at https://github.com/xmed-lab/DiffCMR.
Map-assisted TDOA Localization Enhancement Based On CNN
Nov 09, 2023Abstract:For signal processing related to localization technologies, non line of sight (NLOS) multipaths have great impact over the localization error level. This study proposes a localization correction method based on convolution neural network (CNN) that extracts obstacles' features from maps to predict the localization errors caused by NLOS effects. A novel compensation scheme is developed and structured around the localization error in terms of distance and azimuth angle predicted by the CNN. Four prediction tasks are executed over different building distributions within the maps for typical urban scenario, resulting in CNN models with high prediction accuracy. Finally, a thorough comparison of the accuracy performance between the time difference of arrival (TDOA) localization algorithm and the results after the error compensation reveals that, generally, the CNN prediction approach demonstrates a great localization error correction performance. It can be observed that the powerful feature extraction capability of CNN can be exploited by processing surrounding maps to predict localization error distribution, which has great potential in further enhancement of TDOA performance under challenging scenarios with rich multi-path propagation.
Reconfigurable Intelligent Surface & Edge -- An Introduction of an EM manipulation structure on obstacles' edge
Nov 03, 2023



Abstract:Reconfigurable Intelligent Surface (RIS) or metasurface is one of the important enabling technologies in mobile cellular networks that can effectively enhance the signal coverage performance in obstructed regions, and it is generally deployed on surfaces different from obstacles to redirect electromagnetic (EM) waves by reflection, or covered on objects' surfaces to manipulate EM waves by refraction. In this paper, Reconfigurable Intelligent Surface & Edge (RISE) is proposed to extend RIS' abilities of reflection and refraction over surfaces to diffraction around obstacles' edge for better adaptation to specific coverage scenarios. Based on that, this paper analyzes the performance of several different deployment locations and EM manipulation structure designs for different coverage scenarios. Then a novel EM manipulation structure deployed at the obstacles' edge is proposed to achieve static EM environment modification. Simulations validate the preference of the schemes for different scenarios and the new structure achieves better coverage performance than other typical structures in the static scheme.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Online Easy Example Mining for Weakly-supervised Gland Segmentation from Histology Images
Jun 19, 2022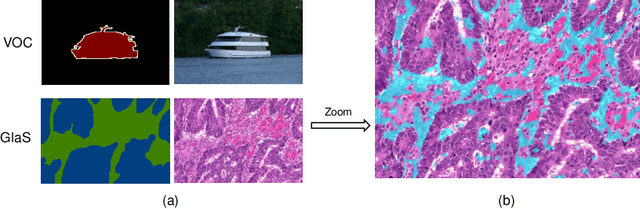
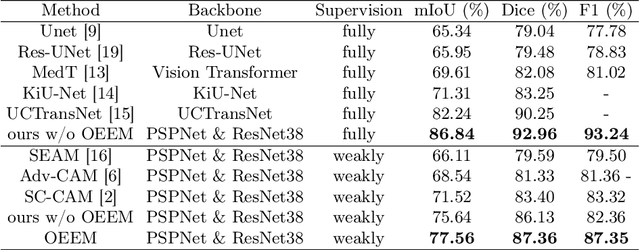
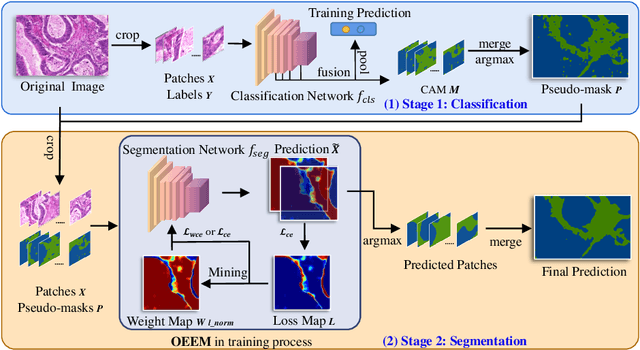
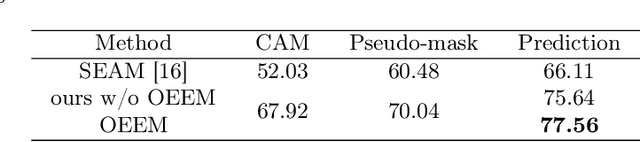
Abstract:Developing an AI-assisted gland segmentation method from histology images is critical for automatic cancer diagnosis and prognosis; however, the high cost of pixel-level annotations hinders its applications to broader diseases. Existing weakly-supervised semantic segmentation methods in computer vision achieve degenerative results for gland segmentation, since the characteristics and problems of glandular datasets are different from general object datasets. We observe that, unlike natural images, the key problem with histology images is the confusion of classes owning to morphological homogeneity and low color contrast among different tissues. To this end, we propose a novel method Online Easy Example Mining (OEEM) that encourages the network to focus on credible supervision signals rather than noisy signals, therefore mitigating the influence of inevitable false predictions in pseudo-masks. According to the characteristics of glandular datasets, we design a strong framework for gland segmentation. Our results exceed many fully-supervised methods and weakly-supervised methods for gland segmentation over 4.4% and 6.04% at mIoU, respectively. Code is available at https://github.com/xmed-lab/OEEM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge