Yiwen Zou
Online Easy Example Mining for Weakly-supervised Gland Segmentation from Histology Images
Jun 19, 2022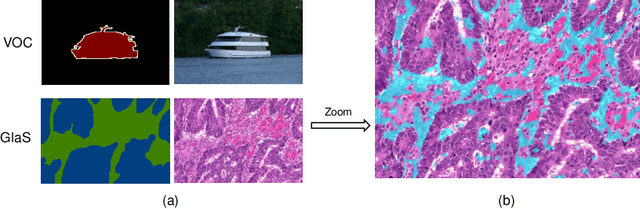
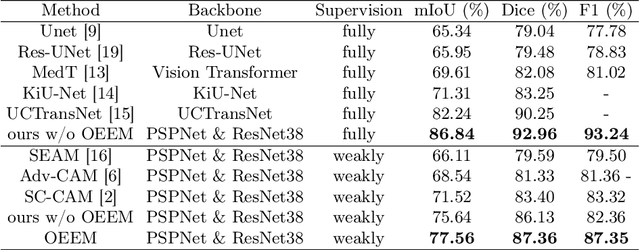
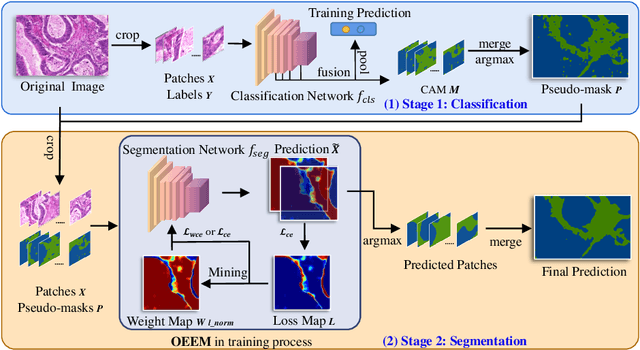
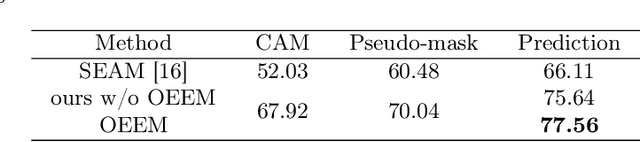
Abstract:Developing an AI-assisted gland segmentation method from histology images is critical for automatic cancer diagnosis and prognosis; however, the high cost of pixel-level annotations hinders its applications to broader diseases. Existing weakly-supervised semantic segmentation methods in computer vision achieve degenerative results for gland segmentation, since the characteristics and problems of glandular datasets are different from general object datasets. We observe that, unlike natural images, the key problem with histology images is the confusion of classes owning to morphological homogeneity and low color contrast among different tissues. To this end, we propose a novel method Online Easy Example Mining (OEEM) that encourages the network to focus on credible supervision signals rather than noisy signals, therefore mitigating the influence of inevitable false predictions in pseudo-masks. According to the characteristics of glandular datasets, we design a strong framework for gland segmentation. Our results exceed many fully-supervised methods and weakly-supervised methods for gland segmentation over 4.4% and 6.04% at mIoU, respectively. Code is available at https://github.com/xmed-lab/OEEM.
WSSS4LUAD: Grand Challenge on Weakly-supervised Tissue Semantic Segmentation for Lung Adenocarcinoma
Apr 14, 2022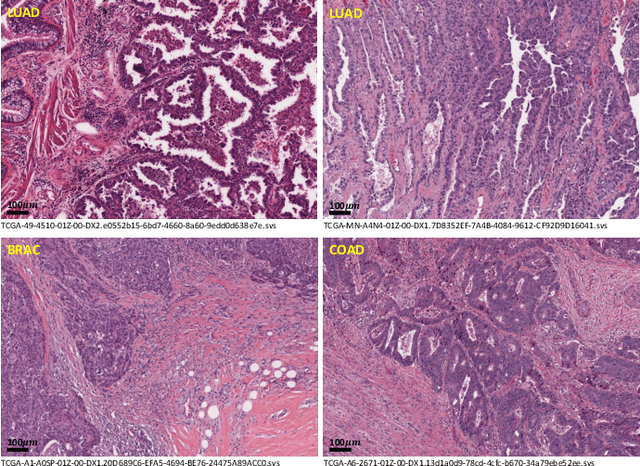
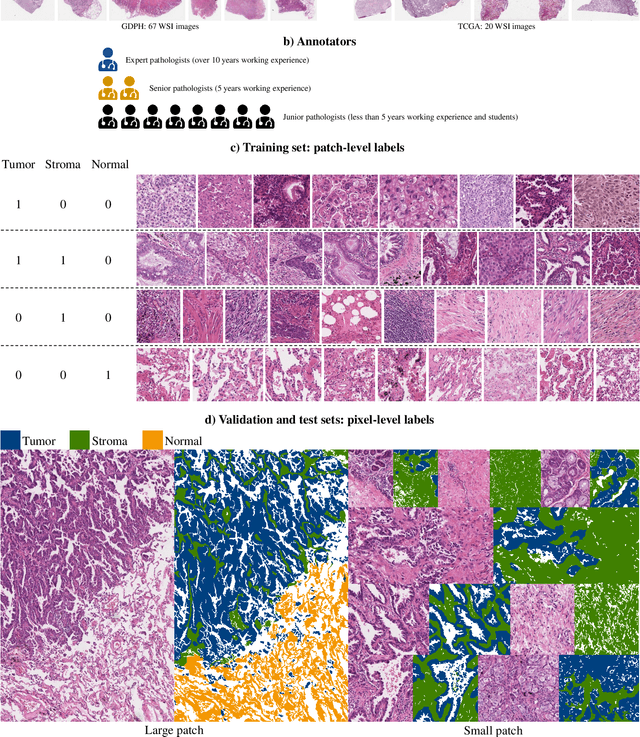
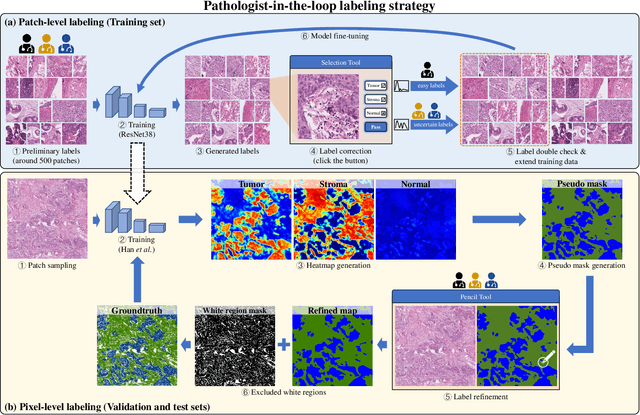
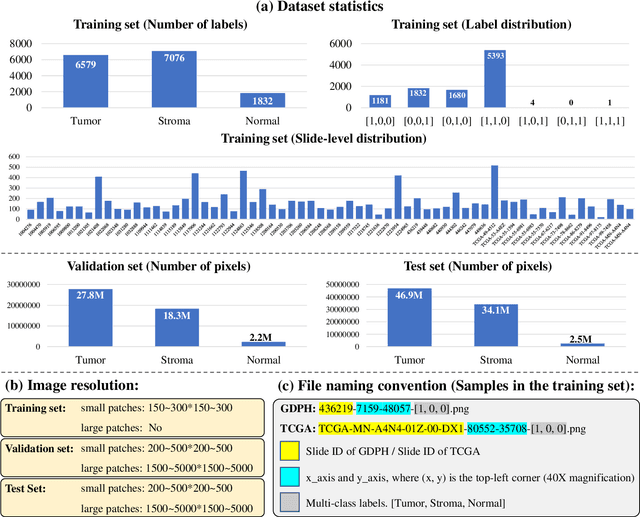
Abstract:Lung cancer is the leading cause of cancer death worldwide, and adenocarcinoma (LUAD) is the most common subtype. Exploiting the potential value of the histopathology images can promote precision medicine in oncology. Tissue segmentation is the basic upstream task of histopathology image analysis. Existing deep learning models have achieved superior segmentation performance but require sufficient pixel-level annotations, which is time-consuming and expensive. To enrich the label resources of LUAD and to alleviate the annotation efforts, we organize this challenge WSSS4LUAD to call for the outstanding weakly-supervised semantic segmentation (WSSS) techniques for histopathology images of LUAD. Participants have to design the algorithm to segment tumor epithelial, tumor-associated stroma and normal tissue with only patch-level labels. This challenge includes 10,091 patch-level annotations (the training set) and over 130 million labeled pixels (the validation and test sets), from 87 WSIs (67 from GDPH, 20 from TCGA). All the labels were generated by a pathologist-in-the-loop pipeline with the help of AI models and checked by the label review board. Among 532 registrations, 28 teams submitted the results in the test phase with over 1,000 submissions. Finally, the first place team achieved mIoU of 0.8413 (tumor: 0.8389, stroma: 0.7931, normal: 0.8919). According to the technical reports of the top-tier teams, CAM is still the most popular approach in WSSS. Cutmix data augmentation has been widely adopted to generate more reliable samples. With the success of this challenge, we believe that WSSS approaches with patch-level annotations can be a complement to the traditional pixel annotations while reducing the annotation efforts. The entire dataset has been released to encourage more researches on computational pathology in LUAD and more novel WSSS techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge