Qixiang Zhang
MOC: Meta-Optimized Classifier for Few-Shot Whole Slide Image Classification
Aug 13, 2025Abstract:Recent advances in histopathology vision-language foundation models (VLFMs) have shown promise in addressing data scarcity for whole slide image (WSI) classification via zero-shot adaptation. However, these methods remain outperformed by conventional multiple instance learning (MIL) approaches trained on large datasets, motivating recent efforts to enhance VLFM-based WSI classification through fewshot learning paradigms. While existing few-shot methods improve diagnostic accuracy with limited annotations, their reliance on conventional classifier designs introduces critical vulnerabilities to data scarcity. To address this problem, we propose a Meta-Optimized Classifier (MOC) comprising two core components: (1) a meta-learner that automatically optimizes a classifier configuration from a mixture of candidate classifiers and (2) a classifier bank housing diverse candidate classifiers to enable a holistic pathological interpretation. Extensive experiments demonstrate that MOC outperforms prior arts in multiple few-shot benchmarks. Notably, on the TCGA-NSCLC benchmark, MOC improves AUC by 10.4% over the state-of-the-art few-shot VLFM-based methods, with gains up to 26.25% under 1-shot conditions, offering a critical advancement for clinical deployments where diagnostic training data is severely limited. Code is available at https://github.com/xmed-lab/MOC.
UniEval: Unified Holistic Evaluation for Unified Multimodal Understanding and Generation
May 15, 2025Abstract:The emergence of unified multimodal understanding and generation models is rapidly attracting attention because of their ability to enhance instruction-following capabilities while minimizing model redundancy. However, there is a lack of a unified evaluation framework for these models, which would enable an elegant, simplified, and overall evaluation. Current models conduct evaluations on multiple task-specific benchmarks, but there are significant limitations, such as the lack of overall results, errors from extra evaluation models, reliance on extensive labeled images, benchmarks that lack diversity, and metrics with limited capacity for instruction-following evaluation. To tackle these challenges, we introduce UniEval, the first evaluation framework designed for unified multimodal models without extra models, images, or annotations. This facilitates a simplified and unified evaluation process. The UniEval framework contains a holistic benchmark, UniBench (supports both unified and visual generation models), along with the corresponding UniScore metric. UniBench includes 81 fine-grained tags contributing to high diversity. Experimental results indicate that UniBench is more challenging than existing benchmarks, and UniScore aligns closely with human evaluations, surpassing current metrics. Moreover, we extensively evaluated SoTA unified and visual generation models, uncovering new insights into Univeral's unique values.
Multi-Modal Explainable Medical AI Assistant for Trustworthy Human-AI Collaboration
May 11, 2025Abstract:Generalist Medical AI (GMAI) systems have demonstrated expert-level performance in biomedical perception tasks, yet their clinical utility remains limited by inadequate multi-modal explainability and suboptimal prognostic capabilities. Here, we present XMedGPT, a clinician-centric, multi-modal AI assistant that integrates textual and visual interpretability to support transparent and trustworthy medical decision-making. XMedGPT not only produces accurate diagnostic and descriptive outputs, but also grounds referenced anatomical sites within medical images, bridging critical gaps in interpretability and enhancing clinician usability. To support real-world deployment, we introduce a reliability indexing mechanism that quantifies uncertainty through consistency-based assessment via interactive question-answering. We validate XMedGPT across four pillars: multi-modal interpretability, uncertainty quantification, and prognostic modeling, and rigorous benchmarking. The model achieves an IoU of 0.703 across 141 anatomical regions, and a Kendall's tau-b of 0.479, demonstrating strong alignment between visual rationales and clinical outcomes. For uncertainty estimation, it attains an AUC of 0.862 on visual question answering and 0.764 on radiology report generation. In survival and recurrence prediction for lung and glioma cancers, it surpasses prior leading models by 26.9%, and outperforms GPT-4o by 25.0%. Rigorous benchmarking across 347 datasets covers 40 imaging modalities and external validation spans 4 anatomical systems confirming exceptional generalizability, with performance gains surpassing existing GMAI by 20.7% for in-domain evaluation and 16.7% on 11,530 in-house data evaluation. Together, XMedGPT represents a significant leap forward in clinician-centric AI integration, offering trustworthy and scalable support for diverse healthcare applications.
Reinforced Correlation Between Vision and Language for Precise Medical AI Assistant
May 06, 2025Abstract:Medical AI assistants support doctors in disease diagnosis, medical image analysis, and report generation. However, they still face significant challenges in clinical use, including limited accuracy with multimodal content and insufficient validation in real-world settings. We propose RCMed, a full-stack AI assistant that improves multimodal alignment in both input and output, enabling precise anatomical delineation, accurate localization, and reliable diagnosis through hierarchical vision-language grounding. A self-reinforcing correlation mechanism allows visual features to inform language context, while language semantics guide pixel-wise attention, forming a closed loop that refines both modalities. This correlation is enhanced by a color region description strategy, translating anatomical structures into semantically rich text to learn shape-location-text relationships across scales. Trained on 20 million image-mask-description triplets, RCMed achieves state-of-the-art precision in contextualizing irregular lesions and subtle anatomical boundaries, excelling in 165 clinical tasks across 9 modalities. It achieved a 23.5% relative improvement in cell segmentation from microscopy images over prior methods. RCMed's strong vision-language alignment enables exceptional generalization, with state-of-the-art performance in external validation across 20 clinically significant cancer types, including novel tasks. This work demonstrates how integrated multimodal models capture fine-grained patterns, enabling human-level interpretation in complex scenarios and advancing human-centric AI healthcare.
Neurons: Emulating the Human Visual Cortex Improves Fidelity and Interpretability in fMRI-to-Video Reconstruction
Mar 14, 2025


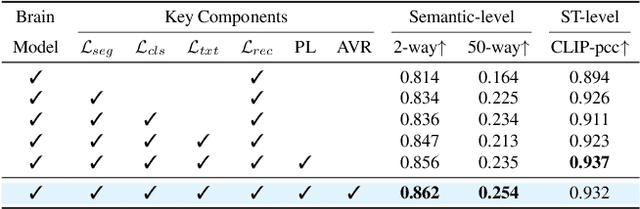
Abstract:Decoding visual stimuli from neural activity is essential for understanding the human brain. While fMRI methods have successfully reconstructed static images, fMRI-to-video reconstruction faces challenges due to the need for capturing spatiotemporal dynamics like motion and scene transitions. Recent approaches have improved semantic and perceptual alignment but struggle to integrate coarse fMRI data with detailed visual features. Inspired by the hierarchical organization of the visual system, we propose NEURONS, a novel framework that decouples learning into four correlated sub-tasks: key object segmentation, concept recognition, scene description, and blurry video reconstruction. This approach simulates the visual cortex's functional specialization, allowing the model to capture diverse video content. In the inference stage, NEURONS generates robust conditioning signals for a pre-trained text-to-video diffusion model to reconstruct the videos. Extensive experiments demonstrate that NEURONS outperforms state-of-the-art baselines, achieving solid improvements in video consistency (26.6%) and semantic-level accuracy (19.1%). Notably, NEURONS shows a strong functional correlation with the visual cortex, highlighting its potential for brain-computer interfaces and clinical applications. Code and model weights will be available at: https://github.com/xmed-lab/NEURONS.
S&D Messenger: Exchanging Semantic and Domain Knowledge for Generic Semi-Supervised Medical Image Segmentation
Jul 10, 2024Abstract:Semi-supervised medical image segmentation (SSMIS) has emerged as a promising solution to tackle the challenges of time-consuming manual labeling in the medical field. However, in practical scenarios, there are often domain variations within the datasets, leading to derivative scenarios like semi-supervised medical domain generalization (Semi-MDG) and unsupervised medical domain adaptation (UMDA). In this paper, we aim to develop a generic framework that masters all three tasks. We notice a critical shared challenge across three scenarios: the explicit semantic knowledge for segmentation performance and rich domain knowledge for generalizability exclusively exist in the labeled set and unlabeled set respectively. Such discrepancy hinders existing methods from effectively comprehending both types of knowledge under semi-supervised settings. To tackle this challenge, we develop a Semantic & Domain Knowledge Messenger (S&D Messenger) which facilitates direct knowledge delivery between the labeled and unlabeled set, and thus allowing the model to comprehend both of them in each individual learning flow. Equipped with our S&D Messenger, a naive pseudo-labeling method can achieve huge improvement on six benchmark datasets for SSMIS (+7.5%), UMDA (+5.6%), and Semi-MDG tasks (+1.14%), compared with state-of-the-art methods designed for specific tasks.
AllSpark: Reborn Labeled Features from Unlabeled in Transformer for Semi-Supervised Semantic Segmentation
Mar 14, 2024Abstract:Semi-supervised semantic segmentation (SSSS) has been proposed to alleviate the burden of time-consuming pixel-level manual labeling, which leverages limited labeled data along with larger amounts of unlabeled data. Current state-of-the-art methods train the labeled data with ground truths and unlabeled data with pseudo labels. However, the two training flows are separate, which allows labeled data to dominate the training process, resulting in low-quality pseudo labels and, consequently, sub-optimal results. To alleviate this issue, we present AllSpark, which reborns the labeled features from unlabeled ones with the channel-wise cross-attention mechanism. We further introduce a Semantic Memory along with a Channel Semantic Grouping strategy to ensure that unlabeled features adequately represent labeled features. The AllSpark shed new light on the architecture level designs of SSSS rather than framework level, which avoids increasingly complicated training pipeline designs. It can also be regarded as a flexible bottleneck module that can be seamlessly integrated into a general transformer-based segmentation model. The proposed AllSpark outperforms existing methods across all evaluation protocols on Pascal, Cityscapes and COCO benchmarks without bells-and-whistles. Code and model weights are available at: https://github.com/xmed-lab/AllSpark.
Morphology-inspired Unsupervised Gland Segmentation via Selective Semantic Grouping
Jul 22, 2023Abstract:Designing deep learning algorithms for gland segmentation is crucial for automatic cancer diagnosis and prognosis, yet the expensive annotation cost hinders the development and application of this technology. In this paper, we make a first attempt to explore a deep learning method for unsupervised gland segmentation, where no manual annotations are required. Existing unsupervised semantic segmentation methods encounter a huge challenge on gland images: They either over-segment a gland into many fractions or under-segment the gland regions by confusing many of them with the background. To overcome this challenge, our key insight is to introduce an empirical cue about gland morphology as extra knowledge to guide the segmentation process. To this end, we propose a novel Morphology-inspired method via Selective Semantic Grouping. We first leverage the empirical cue to selectively mine out proposals for gland sub-regions with variant appearances. Then, a Morphology-aware Semantic Grouping module is employed to summarize the overall information about the gland by explicitly grouping the semantics of its sub-region proposals. In this way, the final segmentation network could learn comprehensive knowledge about glands and produce well-delineated, complete predictions. We conduct experiments on GlaS dataset and CRAG dataset. Our method exceeds the second-best counterpart over 10.56% at mIOU.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge