Shanshan Song
Multi-Modal Explainable Medical AI Assistant for Trustworthy Human-AI Collaboration
May 11, 2025Abstract:Generalist Medical AI (GMAI) systems have demonstrated expert-level performance in biomedical perception tasks, yet their clinical utility remains limited by inadequate multi-modal explainability and suboptimal prognostic capabilities. Here, we present XMedGPT, a clinician-centric, multi-modal AI assistant that integrates textual and visual interpretability to support transparent and trustworthy medical decision-making. XMedGPT not only produces accurate diagnostic and descriptive outputs, but also grounds referenced anatomical sites within medical images, bridging critical gaps in interpretability and enhancing clinician usability. To support real-world deployment, we introduce a reliability indexing mechanism that quantifies uncertainty through consistency-based assessment via interactive question-answering. We validate XMedGPT across four pillars: multi-modal interpretability, uncertainty quantification, and prognostic modeling, and rigorous benchmarking. The model achieves an IoU of 0.703 across 141 anatomical regions, and a Kendall's tau-b of 0.479, demonstrating strong alignment between visual rationales and clinical outcomes. For uncertainty estimation, it attains an AUC of 0.862 on visual question answering and 0.764 on radiology report generation. In survival and recurrence prediction for lung and glioma cancers, it surpasses prior leading models by 26.9%, and outperforms GPT-4o by 25.0%. Rigorous benchmarking across 347 datasets covers 40 imaging modalities and external validation spans 4 anatomical systems confirming exceptional generalizability, with performance gains surpassing existing GMAI by 20.7% for in-domain evaluation and 16.7% on 11,530 in-house data evaluation. Together, XMedGPT represents a significant leap forward in clinician-centric AI integration, offering trustworthy and scalable support for diverse healthcare applications.
DDaTR: Dynamic Difference-aware Temporal Residual Network for Longitudinal Radiology Report Generation
May 06, 2025Abstract:Radiology Report Generation (RRG) automates the creation of radiology reports from medical imaging, enhancing the efficiency of the reporting process. Longitudinal Radiology Report Generation (LRRG) extends RRG by incorporating the ability to compare current and prior exams, facilitating the tracking of temporal changes in clinical findings. Existing LRRG approaches only extract features from prior and current images using a visual pre-trained encoder, which are then concatenated to generate the final report. However, these methods struggle to effectively capture both spatial and temporal correlations during the feature extraction process. Consequently, the extracted features inadequately capture the information of difference across exams and thus underrepresent the expected progressions, leading to sub-optimal performance in LRRG. To address this, we develop a novel dynamic difference-aware temporal residual network (DDaTR). In DDaTR, we introduce two modules at each stage of the visual encoder to capture multi-level spatial correlations. The Dynamic Feature Alignment Module (DFAM) is designed to align prior features across modalities for the integrity of prior clinical information. Prompted by the enriched prior features, the dynamic difference-aware module (DDAM) captures favorable difference information by identifying relationships across exams. Furthermore, our DDaTR employs the dynamic residual network to unidirectionally transmit longitudinal information, effectively modelling temporal correlations. Extensive experiments demonstrated superior performance over existing methods on three benchmarks, proving its efficacy in both RRG and LRRG tasks.
Med-DANet V2: A Flexible Dynamic Architecture for Efficient Medical Volumetric Segmentation
Oct 28, 2023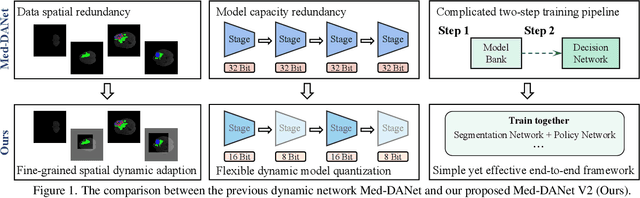
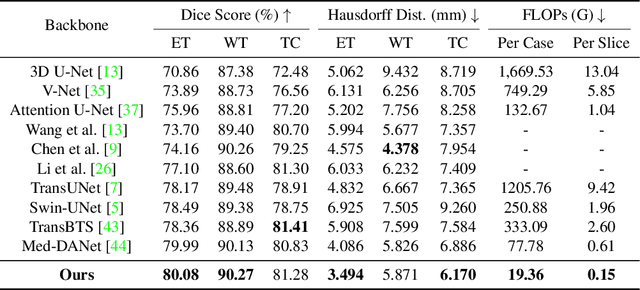
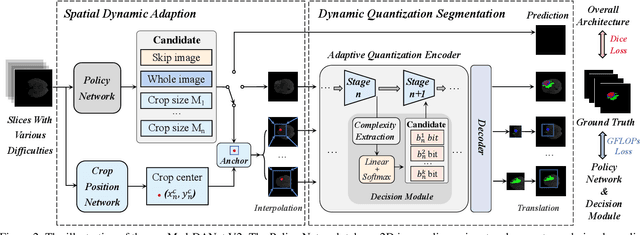
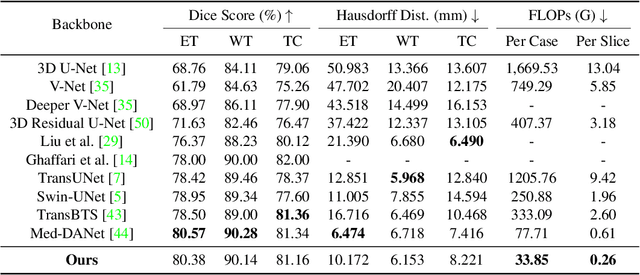
Abstract:Recent works have shown that the computational efficiency of 3D medical image (e.g. CT and MRI) segmentation can be impressively improved by dynamic inference based on slice-wise complexity. As a pioneering work, a dynamic architecture network for medical volumetric segmentation (i.e. Med-DANet) has achieved a favorable accuracy and efficiency trade-off by dynamically selecting a suitable 2D candidate model from the pre-defined model bank for different slices. However, the issues of incomplete data analysis, high training costs, and the two-stage pipeline in Med-DANet require further improvement. To this end, this paper further explores a unified formulation of the dynamic inference framework from the perspective of both the data itself and the model structure. For each slice of the input volume, our proposed method dynamically selects an important foreground region for segmentation based on the policy generated by our Decision Network and Crop Position Network. Besides, we propose to insert a stage-wise quantization selector to the employed segmentation model (e.g. U-Net) for dynamic architecture adapting. Extensive experiments on BraTS 2019 and 2020 show that our method achieves comparable or better performance than previous state-of-the-art methods with much less model complexity. Compared with previous methods Med-DANet and TransBTS with dynamic and static architecture respectively, our framework improves the model efficiency by up to nearly 4.1 and 17.3 times with comparable segmentation results on BraTS 2019.
Wasserstein Generative Regression
Jun 27, 2023Abstract:In this paper, we propose a new and unified approach for nonparametric regression and conditional distribution learning. Our approach simultaneously estimates a regression function and a conditional generator using a generative learning framework, where a conditional generator is a function that can generate samples from a conditional distribution. The main idea is to estimate a conditional generator that satisfies the constraint that it produces a good regression function estimator. We use deep neural networks to model the conditional generator. Our approach can handle problems with multivariate outcomes and covariates, and can be used to construct prediction intervals. We provide theoretical guarantees by deriving non-asymptotic error bounds and the distributional consistency of our approach under suitable assumptions. We also perform numerical experiments with simulated and real data to demonstrate the effectiveness and superiority of our approach over some existing approaches in various scenarios.
FreMAE: Fourier Transform Meets Masked Autoencoders for Medical Image Segmentation
Apr 21, 2023Abstract:The research community has witnessed the powerful potential of self-supervised Masked Image Modeling (MIM), which enables the models capable of learning visual representation from unlabeled data. In this paper, to incorporate both the crucial global structural information and local details for dense prediction tasks, we alter the perspective to the frequency domain and present a new MIM-based framework named FreMAE for self-supervised pre-training for medical image segmentation. Based on the observations that the detailed structural information mainly lies in the high-frequency components and the high-level semantics are abundant in the low-frequency counterparts, we further incorporate multi-stage supervision to guide the representation learning during the pre-training phase. Extensive experiments on three benchmark datasets show the superior advantage of our proposed FreMAE over previous state-of-the-art MIM methods. Compared with various baselines trained from scratch, our FreMAE could consistently bring considerable improvements to the model performance. To the best our knowledge, this is the first attempt towards MIM with Fourier Transform in medical image segmentation.
Med-Tuning: Exploring Parameter-Efficient Transfer Learning for Medical Volumetric Segmentation
Apr 21, 2023Abstract:Deep learning based medical volumetric segmentation methods either train the model from scratch or follow the standard "pre-training then finetuning" paradigm. Although finetuning a well pre-trained model on downstream tasks can harness its representation power, the standard full finetuning is costly in terms of computation and memory footprint. In this paper, we present the first study on parameter-efficient transfer learning for medical volumetric segmentation and propose a novel framework named Med-Tuning based on intra-stage feature enhancement and inter-stage feature interaction. Given a large-scale pre-trained model on 2D natural images, our method can exploit both the multi-scale spatial feature representations and temporal correlations along image slices, which are crucial for accurate medical volumetric segmentation. Extensive experiments on three benchmark datasets (including CT and MRI) show that our method can achieve better results than previous state-of-the-art parameter-efficient transfer learning methods and full finetuning for the segmentation task, with much less tuned parameter costs. Compared to full finetuning, our method reduces the finetuned model parameters by up to 4x, with even better segmentation performance.
MF2-MVQA: A Multi-stage Feature Fusion method for Medical Visual Question Answering
Nov 11, 2022Abstract:There is a key problem in the medical visual question answering task that how to effectively realize the feature fusion of language and medical images with limited datasets. In order to better utilize multi-scale information of medical images, previous methods directly embed the multi-stage visual feature maps as tokens of same size respectively and fuse them with text representation. However, this will cause the confusion of visual features at different stages. To this end, we propose a simple but powerful multi-stage feature fusion method, MF2-MVQA, which stage-wise fuses multi-level visual features with textual semantics. MF2-MVQA achieves the State-Of-The-Art performance on VQA-Med 2019 and VQA-RAD dataset. The results of visualization also verify that our model outperforms previous work.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge