Jiangyun Li
MGD-SAM2: Multi-view Guided Detail-enhanced Segment Anything Model 2 for High-Resolution Class-agnostic Segmentation
Mar 31, 2025Abstract:Segment Anything Models (SAMs), as vision foundation models, have demonstrated remarkable performance across various image analysis tasks. Despite their strong generalization capabilities, SAMs encounter challenges in fine-grained detail segmentation for high-resolution class-independent segmentation (HRCS), due to the limitations in the direct processing of high-resolution inputs and low-resolution mask predictions, and the reliance on accurate manual prompts. To address these limitations, we propose MGD-SAM2 which integrates SAM2 with multi-view feature interaction between a global image and local patches to achieve precise segmentation. MGD-SAM2 incorporates the pre-trained SAM2 with four novel modules: the Multi-view Perception Adapter (MPAdapter), the Multi-view Complementary Enhancement Module (MCEM), the Hierarchical Multi-view Interaction Module (HMIM), and the Detail Refinement Module (DRM). Specifically, we first introduce MPAdapter to adapt the SAM2 encoder for enhanced extraction of local details and global semantics in HRCS images. Then, MCEM and HMIM are proposed to further exploit local texture and global context by aggregating multi-view features within and across multi-scales. Finally, DRM is designed to generate gradually restored high-resolution mask predictions, compensating for the loss of fine-grained details resulting from directly upsampling the low-resolution prediction maps. Experimental results demonstrate the superior performance and strong generalization of our model on multiple high-resolution and normal-resolution datasets. Code will be available at https://github.com/sevenshr/MGD-SAM2.
Referring Remote Sensing Image Segmentation via Bidirectional Alignment Guided Joint Prediction
Feb 12, 2025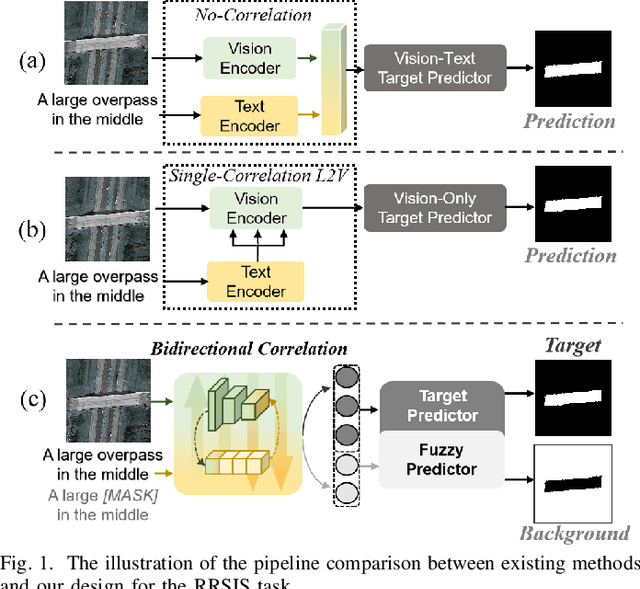



Abstract:Referring Remote Sensing Image Segmentation (RRSIS) is critical for ecological monitoring, urban planning, and disaster management, requiring precise segmentation of objects in remote sensing imagery guided by textual descriptions. This task is uniquely challenging due to the considerable vision-language gap, the high spatial resolution and broad coverage of remote sensing imagery with diverse categories and small targets, and the presence of clustered, unclear targets with blurred edges. To tackle these issues, we propose \ours, a novel framework designed to bridge the vision-language gap, enhance multi-scale feature interaction, and improve fine-grained object differentiation. Specifically, \ours introduces: (1) the Bidirectional Spatial Correlation (BSC) for improved vision-language feature alignment, (2) the Target-Background TwinStream Decoder (T-BTD) for precise distinction between targets and non-targets, and (3) the Dual-Modal Object Learning Strategy (D-MOLS) for robust multimodal feature reconstruction. Extensive experiments on the benchmark datasets RefSegRS and RRSIS-D demonstrate that \ours achieves state-of-the-art performance. Specifically, \ours improves the overall IoU (oIoU) by 3.76 percentage points (80.57) and 1.44 percentage points (79.23) on the two datasets, respectively. Additionally, it outperforms previous methods in the mean IoU (mIoU) by 5.37 percentage points (67.95) and 1.84 percentage points (66.04), effectively addressing the core challenges of RRSIS with enhanced precision and robustness.
Med-DANet V2: A Flexible Dynamic Architecture for Efficient Medical Volumetric Segmentation
Oct 28, 2023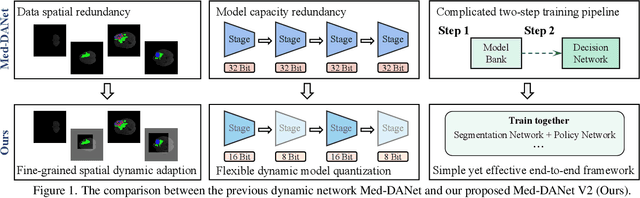
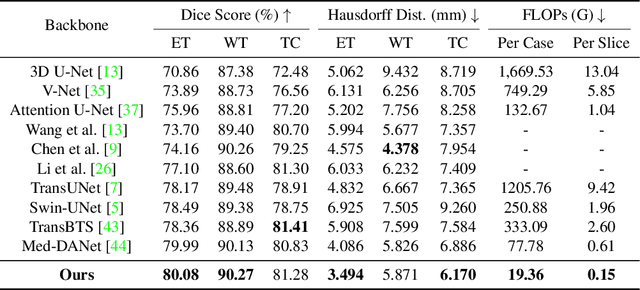
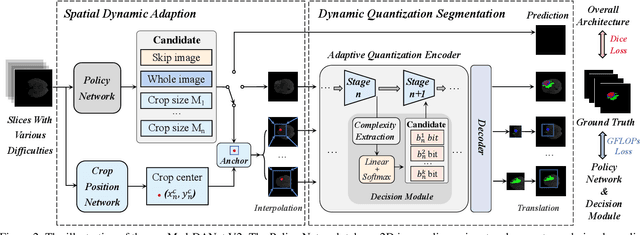
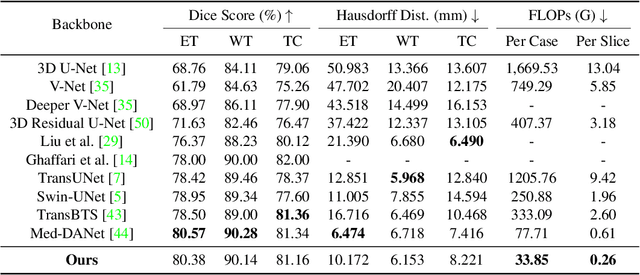
Abstract:Recent works have shown that the computational efficiency of 3D medical image (e.g. CT and MRI) segmentation can be impressively improved by dynamic inference based on slice-wise complexity. As a pioneering work, a dynamic architecture network for medical volumetric segmentation (i.e. Med-DANet) has achieved a favorable accuracy and efficiency trade-off by dynamically selecting a suitable 2D candidate model from the pre-defined model bank for different slices. However, the issues of incomplete data analysis, high training costs, and the two-stage pipeline in Med-DANet require further improvement. To this end, this paper further explores a unified formulation of the dynamic inference framework from the perspective of both the data itself and the model structure. For each slice of the input volume, our proposed method dynamically selects an important foreground region for segmentation based on the policy generated by our Decision Network and Crop Position Network. Besides, we propose to insert a stage-wise quantization selector to the employed segmentation model (e.g. U-Net) for dynamic architecture adapting. Extensive experiments on BraTS 2019 and 2020 show that our method achieves comparable or better performance than previous state-of-the-art methods with much less model complexity. Compared with previous methods Med-DANet and TransBTS with dynamic and static architecture respectively, our framework improves the model efficiency by up to nearly 4.1 and 17.3 times with comparable segmentation results on BraTS 2019.
CM-MaskSD: Cross-Modality Masked Self-Distillation for Referring Image Segmentation
May 22, 2023Abstract:Referring image segmentation (RIS) is a fundamental vision-language task that intends to segment a desired object from an image based on a given natural language expression. Due to the essentially distinct data properties between image and text, most of existing methods either introduce complex designs towards fine-grained vision-language alignment or lack required dense alignment, resulting in scalability issues or mis-segmentation problems such as over- or under-segmentation. To achieve effective and efficient fine-grained feature alignment in the RIS task, we explore the potential of masked multimodal modeling coupled with self-distillation and propose a novel cross-modality masked self-distillation framework named CM-MaskSD, in which our method inherits the transferred knowledge of image-text semantic alignment from CLIP model to realize fine-grained patch-word feature alignment for better segmentation accuracy. Moreover, our CM-MaskSD framework can considerably boost model performance in a nearly parameter-free manner, since it shares weights between the main segmentation branch and the introduced masked self-distillation branches, and solely introduces negligible parameters for coordinating the multimodal features. Comprehensive experiments on three benchmark datasets (i.e. RefCOCO, RefCOCO+, G-Ref) for the RIS task convincingly demonstrate the superiority of our proposed framework over previous state-of-the-art methods.
FreMAE: Fourier Transform Meets Masked Autoencoders for Medical Image Segmentation
Apr 21, 2023Abstract:The research community has witnessed the powerful potential of self-supervised Masked Image Modeling (MIM), which enables the models capable of learning visual representation from unlabeled data. In this paper, to incorporate both the crucial global structural information and local details for dense prediction tasks, we alter the perspective to the frequency domain and present a new MIM-based framework named FreMAE for self-supervised pre-training for medical image segmentation. Based on the observations that the detailed structural information mainly lies in the high-frequency components and the high-level semantics are abundant in the low-frequency counterparts, we further incorporate multi-stage supervision to guide the representation learning during the pre-training phase. Extensive experiments on three benchmark datasets show the superior advantage of our proposed FreMAE over previous state-of-the-art MIM methods. Compared with various baselines trained from scratch, our FreMAE could consistently bring considerable improvements to the model performance. To the best our knowledge, this is the first attempt towards MIM with Fourier Transform in medical image segmentation.
Med-Tuning: Exploring Parameter-Efficient Transfer Learning for Medical Volumetric Segmentation
Apr 21, 2023Abstract:Deep learning based medical volumetric segmentation methods either train the model from scratch or follow the standard "pre-training then finetuning" paradigm. Although finetuning a well pre-trained model on downstream tasks can harness its representation power, the standard full finetuning is costly in terms of computation and memory footprint. In this paper, we present the first study on parameter-efficient transfer learning for medical volumetric segmentation and propose a novel framework named Med-Tuning based on intra-stage feature enhancement and inter-stage feature interaction. Given a large-scale pre-trained model on 2D natural images, our method can exploit both the multi-scale spatial feature representations and temporal correlations along image slices, which are crucial for accurate medical volumetric segmentation. Extensive experiments on three benchmark datasets (including CT and MRI) show that our method can achieve better results than previous state-of-the-art parameter-efficient transfer learning methods and full finetuning for the segmentation task, with much less tuned parameter costs. Compared to full finetuning, our method reduces the finetuned model parameters by up to 4x, with even better segmentation performance.
MF2-MVQA: A Multi-stage Feature Fusion method for Medical Visual Question Answering
Nov 11, 2022Abstract:There is a key problem in the medical visual question answering task that how to effectively realize the feature fusion of language and medical images with limited datasets. In order to better utilize multi-scale information of medical images, previous methods directly embed the multi-stage visual feature maps as tokens of same size respectively and fuse them with text representation. However, this will cause the confusion of visual features at different stages. To this end, we propose a simple but powerful multi-stage feature fusion method, MF2-MVQA, which stage-wise fuses multi-level visual features with textual semantics. MF2-MVQA achieves the State-Of-The-Art performance on VQA-Med 2019 and VQA-RAD dataset. The results of visualization also verify that our model outperforms previous work.
Positive-Negative Equal Contrastive Loss for Semantic Segmentation
Jul 05, 2022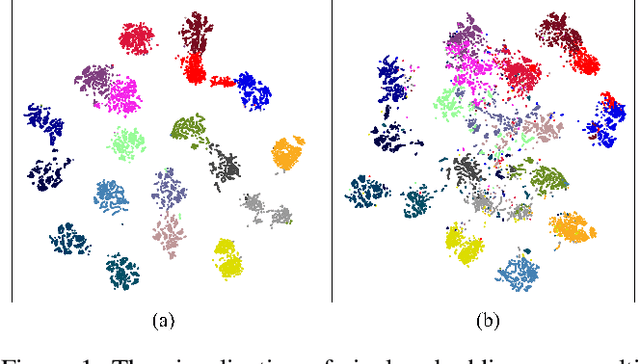
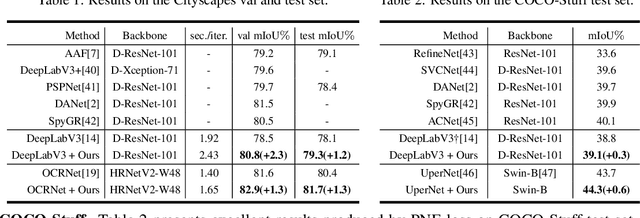
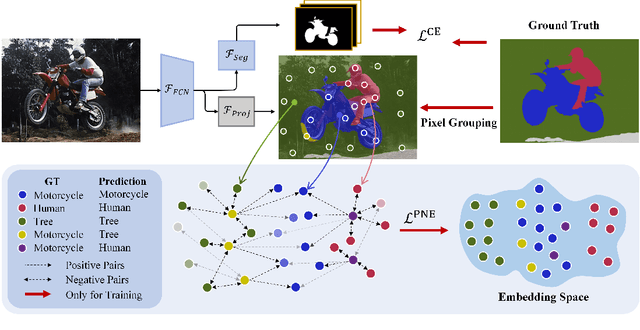
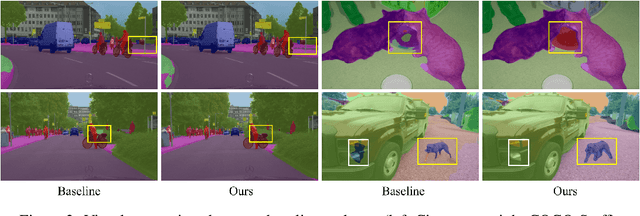
Abstract:The contextual information is critical for various computer vision tasks, previous works commonly design plug-and-play modules and structural losses to effectively extract and aggregate the global context. These methods utilize fine-label to optimize the model but ignore that fine-trained features are also precious training resources, which can introduce preferable distribution to hard pixels (i.e., misclassified pixels). Inspired by contrastive learning in unsupervised paradigm, we apply the contrastive loss in a supervised manner and re-design the loss function to cast off the stereotype of unsupervised learning (e.g., imbalance of positives and negatives, confusion of anchors computing). To this end, we propose Positive-Negative Equal contrastive loss (PNE loss), which increases the latent impact of positive embedding on the anchor and treats the positive as well as negative sample pairs equally. The PNE loss can be directly plugged right into existing semantic segmentation frameworks and leads to excellent performance with neglectable extra computational costs. We utilize a number of classic segmentation methods (e.g., DeepLabV3, OCRNet, UperNet) and backbone (e.g., ResNet, HRNet, Swin Transformer) to conduct comprehensive experiments and achieve state-of-the-art performance on two benchmark datasets (e.g., Cityscapes and COCO-Stuff). Our code will be publicly available soon.
Med-DANet: Dynamic Architecture Network for Efficient Medical Volumetric Segmentation
Jun 14, 2022

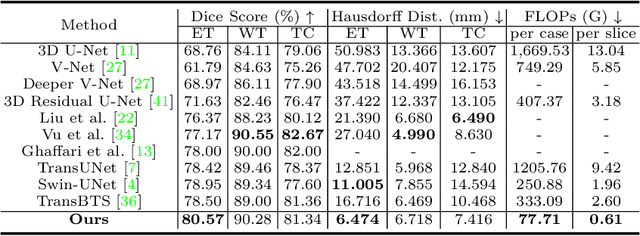
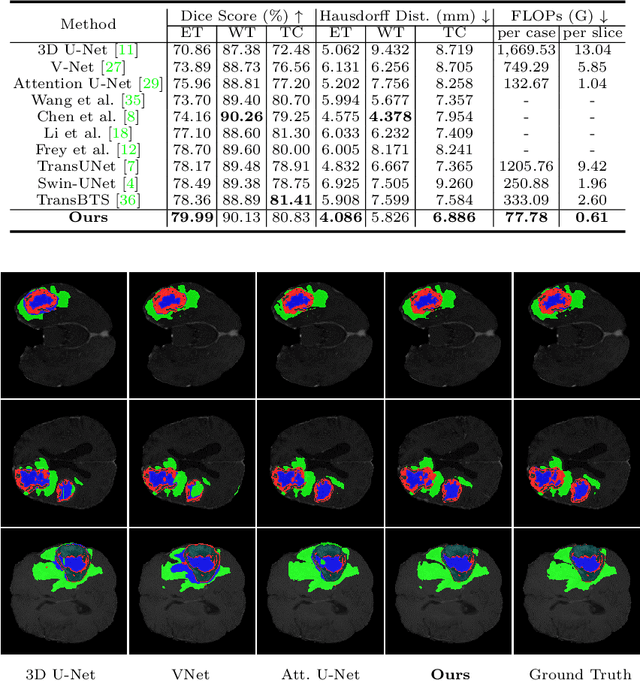
Abstract:For 3D medical image (e.g. CT and MRI) segmentation, the difficulty of segmenting each slice in a clinical case varies greatly. Previous research on volumetric medical image segmentation in a slice-by-slice manner conventionally use the identical 2D deep neural network to segment all the slices of the same case, ignoring the data heterogeneity among image slices. In this paper, we focus on multi-modal 3D MRI brain tumor segmentation and propose a dynamic architecture network named Med-DANet based on adaptive model selection to achieve effective accuracy and efficiency trade-off. For each slice of the input 3D MRI volume, our proposed method learns a slice-specific decision by the Decision Network to dynamically select a suitable model from the predefined Model Bank for the subsequent 2D segmentation task. Extensive experimental results on both BraTS 2019 and 2020 datasets show that our proposed method achieves comparable or better results than previous state-of-the-art methods for 3D MRI brain tumor segmentation with much less model complexity. Compared with the state-of-the-art 3D method TransBTS, the proposed framework improves the model efficiency by up to 3.5x without sacrificing the accuracy. Our code will be publicly available soon.
Attention guided global enhancement and local refinement network for semantic segmentation
Apr 09, 2022
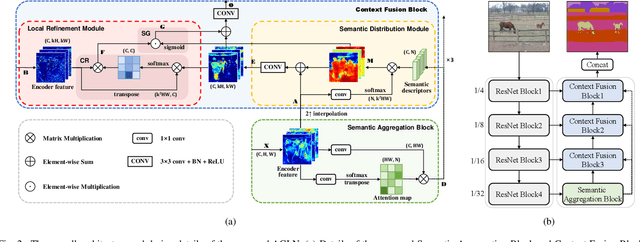
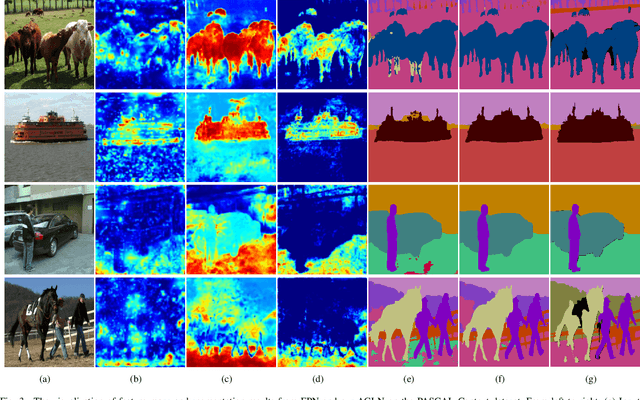
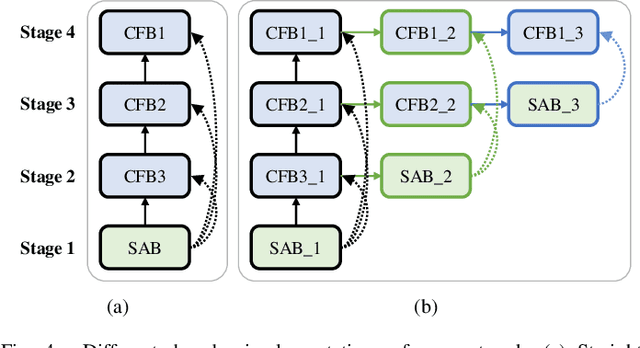
Abstract:The encoder-decoder architecture is widely used as a lightweight semantic segmentation network. However, it struggles with a limited performance compared to a well-designed Dilated-FCN model for two major problems. First, commonly used upsampling methods in the decoder such as interpolation and deconvolution suffer from a local receptive field, unable to encode global contexts. Second, low-level features may bring noises to the network decoder through skip connections for the inadequacy of semantic concepts in early encoder layers. To tackle these challenges, a Global Enhancement Method is proposed to aggregate global information from high-level feature maps and adaptively distribute them to different decoder layers, alleviating the shortage of global contexts in the upsampling process. Besides, a Local Refinement Module is developed by utilizing the decoder features as the semantic guidance to refine the noisy encoder features before the fusion of these two (the decoder features and the encoder features). Then, the two methods are integrated into a Context Fusion Block, and based on that, a novel Attention guided Global enhancement and Local refinement Network (AGLN) is elaborately designed. Extensive experiments on PASCAL Context, ADE20K, and PASCAL VOC 2012 datasets have demonstrated the effectiveness of the proposed approach. In particular, with a vanilla ResNet-101 backbone, AGLN achieves the state-of-the-art result (56.23% mean IoU) on the PASCAL Context dataset. The code is available at https://github.com/zhasen1996/AGLN.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge