Sen Zha
Med-DANet: Dynamic Architecture Network for Efficient Medical Volumetric Segmentation
Jun 14, 2022

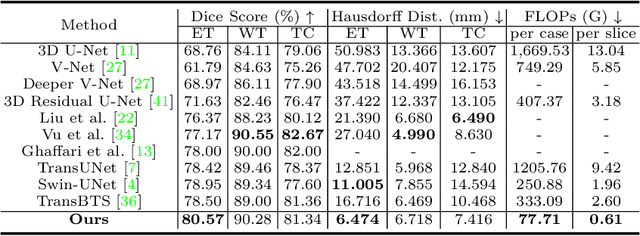
Abstract:For 3D medical image (e.g. CT and MRI) segmentation, the difficulty of segmenting each slice in a clinical case varies greatly. Previous research on volumetric medical image segmentation in a slice-by-slice manner conventionally use the identical 2D deep neural network to segment all the slices of the same case, ignoring the data heterogeneity among image slices. In this paper, we focus on multi-modal 3D MRI brain tumor segmentation and propose a dynamic architecture network named Med-DANet based on adaptive model selection to achieve effective accuracy and efficiency trade-off. For each slice of the input 3D MRI volume, our proposed method learns a slice-specific decision by the Decision Network to dynamically select a suitable model from the predefined Model Bank for the subsequent 2D segmentation task. Extensive experimental results on both BraTS 2019 and 2020 datasets show that our proposed method achieves comparable or better results than previous state-of-the-art methods for 3D MRI brain tumor segmentation with much less model complexity. Compared with the state-of-the-art 3D method TransBTS, the proposed framework improves the model efficiency by up to 3.5x without sacrificing the accuracy. Our code will be publicly available soon.
Attention guided global enhancement and local refinement network for semantic segmentation
Apr 09, 2022
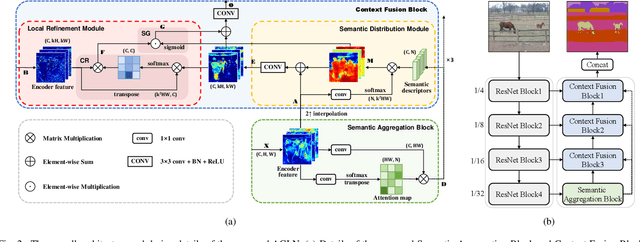
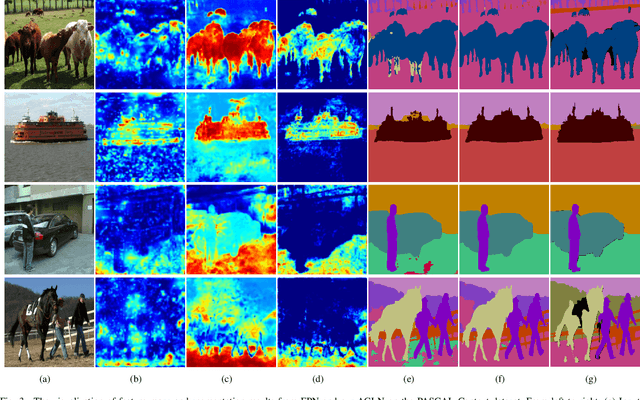
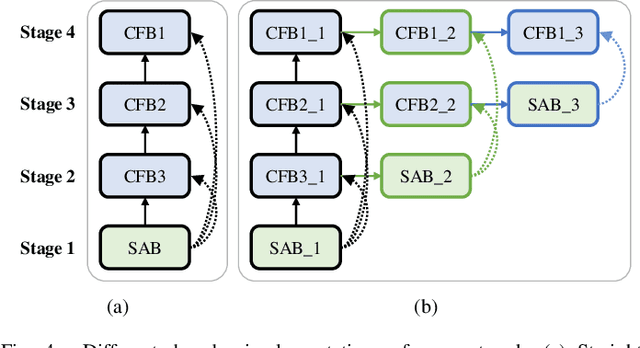
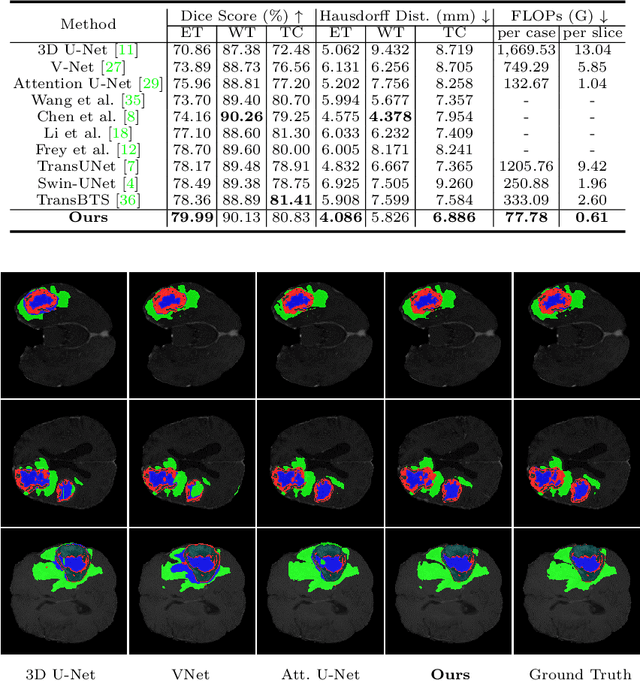
Abstract:The encoder-decoder architecture is widely used as a lightweight semantic segmentation network. However, it struggles with a limited performance compared to a well-designed Dilated-FCN model for two major problems. First, commonly used upsampling methods in the decoder such as interpolation and deconvolution suffer from a local receptive field, unable to encode global contexts. Second, low-level features may bring noises to the network decoder through skip connections for the inadequacy of semantic concepts in early encoder layers. To tackle these challenges, a Global Enhancement Method is proposed to aggregate global information from high-level feature maps and adaptively distribute them to different decoder layers, alleviating the shortage of global contexts in the upsampling process. Besides, a Local Refinement Module is developed by utilizing the decoder features as the semantic guidance to refine the noisy encoder features before the fusion of these two (the decoder features and the encoder features). Then, the two methods are integrated into a Context Fusion Block, and based on that, a novel Attention guided Global enhancement and Local refinement Network (AGLN) is elaborately designed. Extensive experiments on PASCAL Context, ADE20K, and PASCAL VOC 2012 datasets have demonstrated the effectiveness of the proposed approach. In particular, with a vanilla ResNet-101 backbone, AGLN achieves the state-of-the-art result (56.23% mean IoU) on the PASCAL Context dataset. The code is available at https://github.com/zhasen1996/AGLN.
Category Guided Attention Network for Brain Tumor Segmentation in MRI
Mar 29, 2022



Abstract:Objective: Magnetic resonance imaging (MRI) has been widely used for the analysis and diagnosis of brain diseases. Accurate and automatic brain tumor segmentation is of paramount importance for radiation treatment. However, low tissue contrast in tumor regions makes it a challenging task.Approach: We propose a novel segmentation network named Category Guided Attention U-Net (CGA U-Net). In this model, we design a Supervised Attention Module (SAM) based on the attention mechanism, which can capture more accurate and stable long-range dependency in feature maps without introducing much computational cost. Moreover, we propose an intra-class update approach to reconstruct feature maps by aggregating pixels of the same category. Main results: Experimental results on the BraTS 2019 datasets show that the proposed method outperformers the state-of-the-art algorithms in both segmentation performance and computational complexity. Significance: The CGA U-Net can effectively capture the global semantic information in the MRI image by using the SAM module, while significantly reducing the computational cost. Code is available at https://github.com/delugewalker/CGA-U-Net.
TransBTSV2: Wider Instead of Deeper Transformer for Medical Image Segmentation
Jan 30, 2022



Abstract:Transformer, benefiting from global (long-range) information modeling using self-attention mechanism, has been successful in natural language processing and computer vision recently. Convolutional Neural Networks, capable of capturing local features, are unable to model explicit long-distance dependencies from global feature space. However, both local and global features are crucial for dense prediction tasks, especially for 3D medical image segmentation. In this paper, we exploit Transformer in 3D CNN for 3D medical image volumetric segmentation and propose a novel network named TransBTSV2 based on the encoder-decoder structure. Different from our original TransBTS, the proposed TransBTSV2 is not limited to brain tumor segmentation (BTS) but focuses on general medical image segmentation, providing a strong and efficient 3D baseline for volumetric segmentation of medical images. As a hybrid CNN-Transformer architecture, TransBTSV2 can achieve accurate segmentation of medical images without any pre-training. With the proposed insight to redesign the internal structure of Transformer and the introduced Deformable Bottleneck Module, a highly efficient architecture is achieved with superior performance. Extensive experimental results on four medical image datasets (BraTS 2019, BraTS 2020, LiTS 2017 and KiTS 2019) demonstrate that TransBTSV2 achieves comparable or better results as compared to the state-of-the-art methods for the segmentation of brain tumor, liver tumor as well as kidney tumor. Code is available at https://github.com/Wenxuan-1119/TransBTS.
TransBTS: Multimodal Brain Tumor Segmentation Using Transformer
Mar 07, 2021



Abstract:Transformer, which can benefit from global (long-range) information modeling using self-attention mechanisms, has been successful in natural language processing and 2D image classification recently. However, both local and global features are crucial for dense prediction tasks, especially for 3D medical image segmentation. In this paper, we for the first time exploit Transformer in 3D CNN for MRI Brain Tumor Segmentation and propose a novel network named TransBTS based on the encoder-decoder structure. To capture the local 3D context information, the encoder first utilizes 3D CNN to extract the volumetric spatial feature maps. Meanwhile, the feature maps are reformed elaborately for tokens that are fed into Transformer for global feature modeling. The decoder leverages the features embedded by Transformer and performs progressive upsampling to predict the detailed segmentation map. Experimental results on the BraTS 2019 dataset show that TransBTS outperforms state-of-the-art methods for brain tumor segmentation on 3D MRI scans. Code is available at https://github.com/Wenxuan-1119/TransBTS
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge