Virginia Newcombe
Continuous Online Adaptation Driven by User Interaction for Medical Image Segmentation
Mar 09, 2025Abstract:Interactive segmentation models use real-time user interactions, such as mouse clicks, as extra inputs to dynamically refine the model predictions. After model deployment, user corrections of model predictions could be used to adapt the model to the post-deployment data distribution, countering distribution-shift and enhancing reliability. Motivated by this, we introduce an online adaptation framework that enables an interactive segmentation model to continuously learn from user interaction and improve its performance on new data distributions, as it processes a sequence of test images. We introduce the Gaussian Point Loss function to train the model how to leverage user clicks, along with a two-stage online optimization method that adapts the model using the corrected predictions generated via user interactions. We demonstrate that this simple and therefore practical approach is very effective. Experiments on 5 fundus and 4 brain MRI databases demonstrate that our method outperforms existing approaches under various data distribution shifts, including segmentation of image modalities and pathologies not seen during training.
Feasibility and benefits of joint learning from MRI databases with different brain diseases and modalities for segmentation
May 28, 2024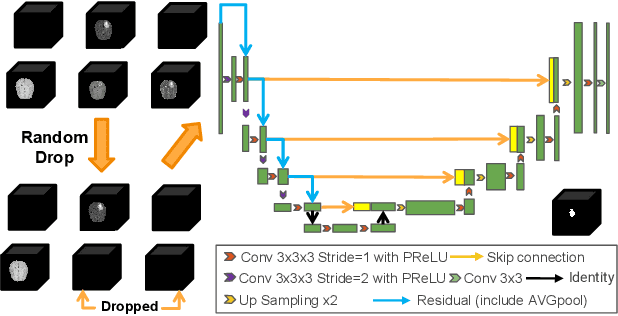
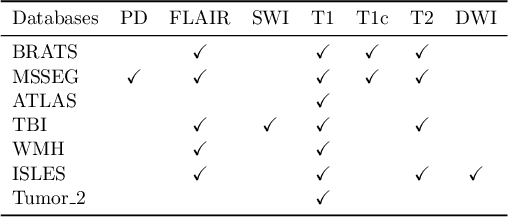
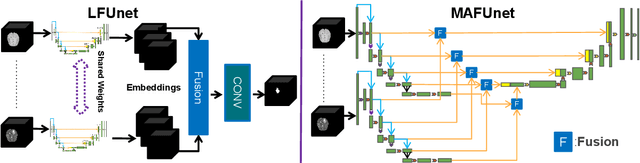

Abstract:Models for segmentation of brain lesions in multi-modal MRI are commonly trained for a specific pathology using a single database with a predefined set of MRI modalities, determined by a protocol for the specific disease. This work explores the following open questions: Is it feasible to train a model using multiple databases that contain varying sets of MRI modalities and annotations for different brain pathologies? Will this joint learning benefit performance on the sets of modalities and pathologies available during training? Will it enable analysis of new databases with different sets of modalities and pathologies? We develop and compare different methods and show that promising results can be achieved with appropriate, simple and practical alterations to the model and training framework. We experiment with 7 databases containing 5 types of brain pathologies and different sets of MRI modalities. Results demonstrate, for the first time, that joint training on multi-modal MRI databases with different brain pathologies and sets of modalities is feasible and offers practical benefits. It enables a single model to segment pathologies encountered during training in diverse sets of modalities, while facilitating segmentation of new types of pathologies such as via follow-up fine-tuning. The insights this study provides into the potential and limitations of this paradigm should prove useful for guiding future advances in the direction. Code and pretrained models: https://github.com/WenTXuL/MultiUnet
* Accepted to MIDL 2024
Linking Symptom Inventories using Semantic Textual Similarity
Sep 08, 2023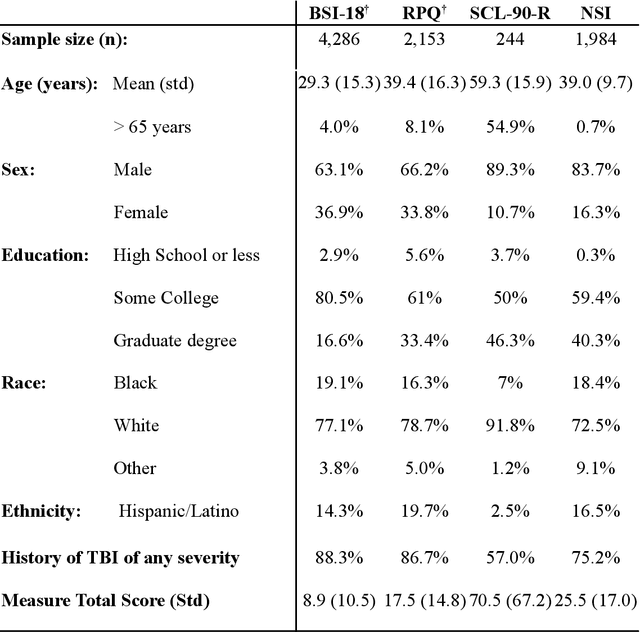
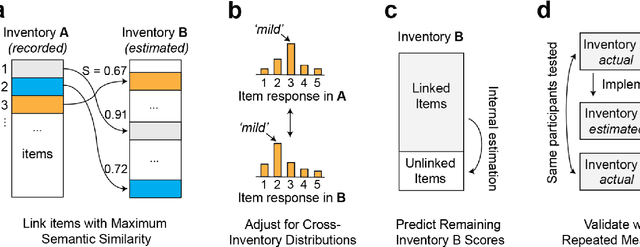
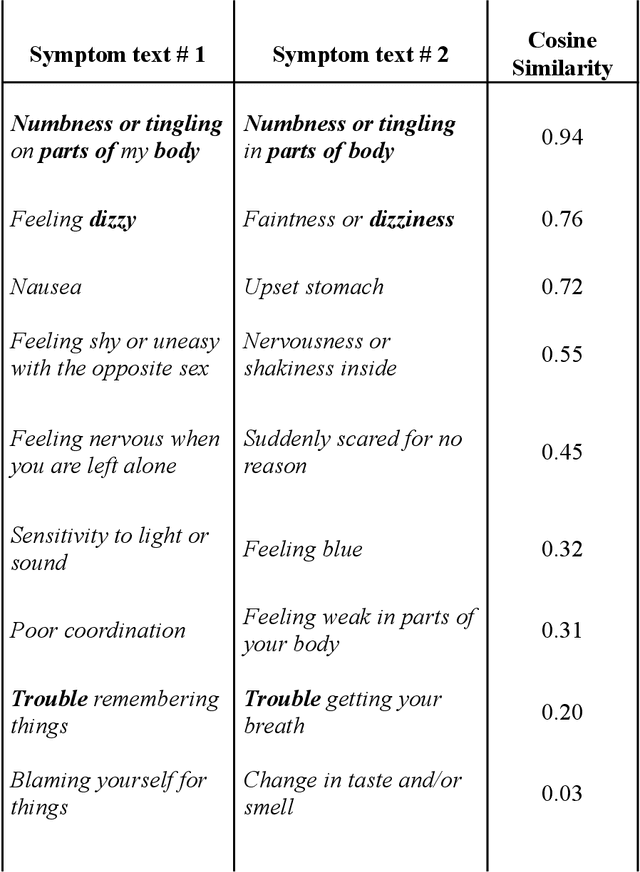
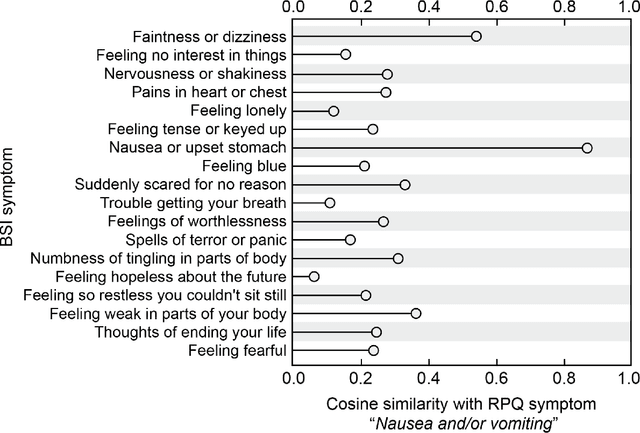
Abstract:An extensive library of symptom inventories has been developed over time to measure clinical symptoms, but this variety has led to several long standing issues. Most notably, results drawn from different settings and studies are not comparable, which limits reproducibility. Here, we present an artificial intelligence (AI) approach using semantic textual similarity (STS) to link symptoms and scores across previously incongruous symptom inventories. We tested the ability of four pre-trained STS models to screen thousands of symptom description pairs for related content - a challenging task typically requiring expert panels. Models were tasked to predict symptom severity across four different inventories for 6,607 participants drawn from 16 international data sources. The STS approach achieved 74.8% accuracy across five tasks, outperforming other models tested. This work suggests that incorporating contextual, semantic information can assist expert decision-making processes, yielding gains for both general and disease-specific clinical assessment.
Cranial Implant Design via Virtual Craniectomy with Shape Priors
Sep 29, 2020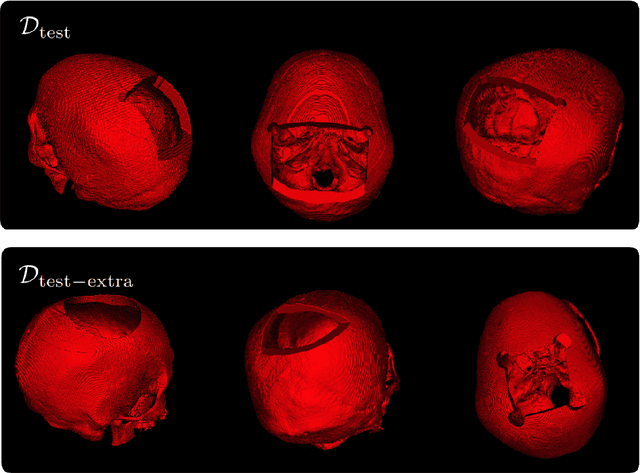
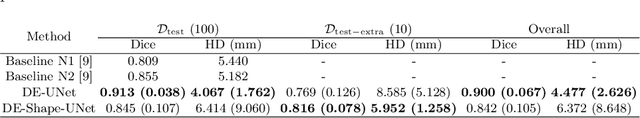
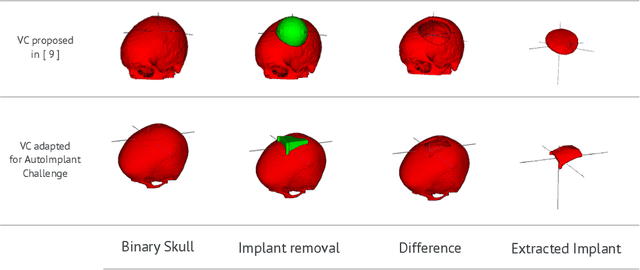
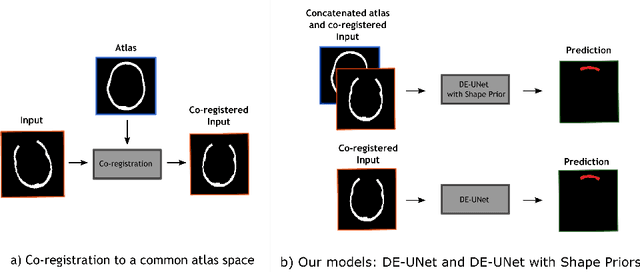
Abstract:Cranial implant design is a challenging task, whose accuracy is crucial in the context of cranioplasty procedures. This task is usually performed manually by experts using computer-assisted design software. In this work, we propose and evaluate alternative automatic deep learning models for cranial implant reconstruction from CT images. The models are trained and evaluated using the database released by the AutoImplant challenge, and compared to a baseline implemented by the organizers. We employ a simulated virtual craniectomy to train our models using complete skulls, and compare two different approaches trained with this procedure. The first one is a direct estimation method based on the UNet architecture. The second method incorporates shape priors to increase the robustness when dealing with out-of-distribution implant shapes. Our direct estimation method outperforms the baselines provided by the organizers, while the model with shape priors shows superior performance when dealing with out-of-distribution cases. Overall, our methods show promising results in the difficult task of cranial implant design.
Self-supervised Skull Reconstruction in Brain CT Images with Decompressive Craniectomy
Jul 11, 2020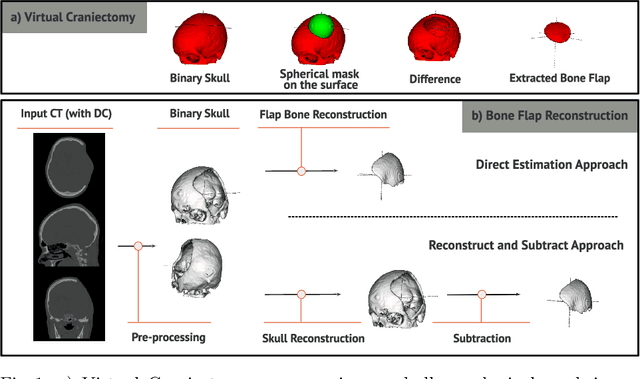
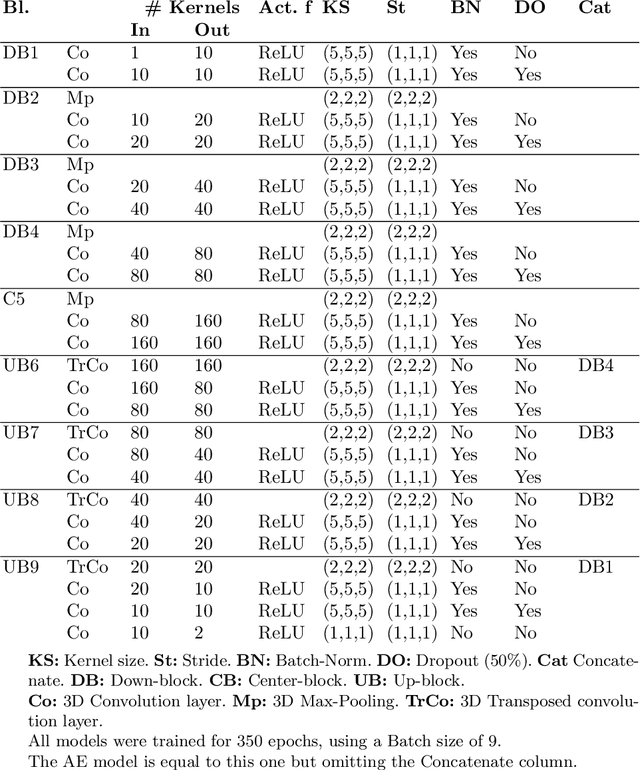
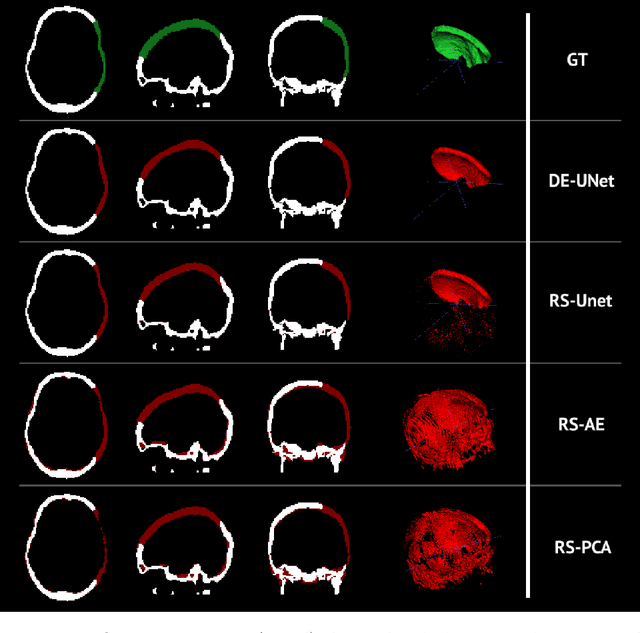
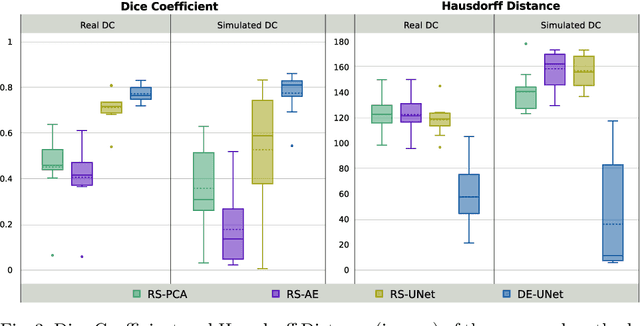
Abstract:Decompressive craniectomy (DC) is a common surgical procedure consisting of the removal of a portion of the skull that is performed after incidents such as stroke, traumatic brain injury (TBI) or other events that could result in acute subdural hemorrhage and/or increasing intracranial pressure. In these cases, CT scans are obtained to diagnose and assess injuries, or guide a certain therapy and intervention. We propose a deep learning based method to reconstruct the skull defect removed during DC performed after TBI from post-operative CT images. This reconstruction is useful in multiple scenarios, e.g. to support the creation of cranioplasty plates, accurate measurements of bone flap volume and total intracranial volume, important for studies that aim to relate later atrophy to patient outcome. We propose and compare alternative self-supervised methods where an encoder-decoder convolutional neural network (CNN) estimates the missing bone flap on post-operative CTs. The self-supervised learning strategy only requires images with complete skulls and avoids the need for annotated DC images. For evaluation, we employ real and simulated images with DC, comparing the results with other state-of-the-art approaches. The experiments show that the proposed model outperforms current manual methods, enabling reconstruction even in highly challenging cases where big skull defects have been removed during surgery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge