Wentian Xu
Modality-Agnostic Input Channels Enable Segmentation of Brain lesions in Multimodal MRI with Sequences Unavailable During Training
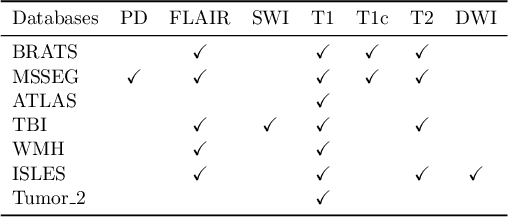
Sep 11, 2025Abstract:Segmentation models are important tools for the detection and analysis of lesions in brain MRI. Depending on the type of brain pathology that is imaged, MRI scanners can acquire multiple, different image modalities (contrasts). Most segmentation models for multimodal brain MRI are restricted to fixed modalities and cannot effectively process new ones at inference. Some models generalize to unseen modalities but may lose discriminative modality-specific information. This work aims to develop a model that can perform inference on data that contain image modalities unseen during training, previously seen modalities, and heterogeneous combinations of both, thus allowing a user to utilize any available imaging modalities. We demonstrate this is possible with a simple, thus practical alteration to the U-net architecture, by integrating a modality-agnostic input channel or pathway, alongside modality-specific input channels. To train this modality-agnostic component, we develop an image augmentation scheme that synthesizes artificial MRI modalities. Augmentations differentially alter the appearance of pathological and healthy brain tissue to create artificial contrasts between them while maintaining realistic anatomical integrity. We evaluate the method using 8 MRI databases that include 5 types of pathologies (stroke, tumours, traumatic brain injury, multiple sclerosis and white matter hyperintensities) and 8 modalities (T1, T1+contrast, T2, PD, SWI, DWI, ADC and FLAIR). The results demonstrate that the approach preserves the ability to effectively process MRI modalities encountered during training, while being able to process new, unseen modalities to improve its segmentation. Project code: https://github.com/Anthony-P-Addison/AGN-MOD-SEG
Specialised or Generic? Tokenization Choices for Radiology Language Models
Aug 13, 2025Abstract:The vocabulary used by language models (LM) - defined by the tokenizer - plays a key role in text generation quality. However, its impact remains under-explored in radiology. In this work, we address this gap by systematically comparing general, medical, and domain-specific tokenizers on the task of radiology report summarisation across three imaging modalities. We also investigate scenarios with and without LM pre-training on PubMed abstracts. Our findings demonstrate that medical and domain-specific vocabularies outperformed widely used natural language alternatives when models are trained from scratch. Pre-training partially mitigates performance differences between tokenizers, whilst the domain-specific tokenizers achieve the most favourable results. Domain-specific tokenizers also reduce memory requirements due to smaller vocabularies and shorter sequences. These results demonstrate that adapting the vocabulary of LMs to the clinical domain provides practical benefits, including improved performance and reduced computational demands, making such models more accessible and effective for both research and real-world healthcare settings.
IterMask3D: Unsupervised Anomaly Detection and Segmentation with Test-Time Iterative Mask Refinement in 3D Brain MR
Apr 07, 2025Abstract:Unsupervised anomaly detection and segmentation methods train a model to learn the training distribution as 'normal'. In the testing phase, they identify patterns that deviate from this normal distribution as 'anomalies'. To learn the `normal' distribution, prevailing methods corrupt the images and train a model to reconstruct them. During testing, the model attempts to reconstruct corrupted inputs based on the learned 'normal' distribution. Deviations from this distribution lead to high reconstruction errors, which indicate potential anomalies. However, corrupting an input image inevitably causes information loss even in normal regions, leading to suboptimal reconstruction and an increased risk of false positives. To alleviate this, we propose IterMask3D, an iterative spatial mask-refining strategy designed for 3D brain MRI. We iteratively spatially mask areas of the image as corruption and reconstruct them, then shrink the mask based on reconstruction error. This process iteratively unmasks 'normal' areas to the model, whose information further guides reconstruction of 'normal' patterns under the mask to be reconstructed accurately, reducing false positives. In addition, to achieve better reconstruction performance, we also propose using high-frequency image content as additional structural information to guide the reconstruction of the masked area. Extensive experiments on the detection of both synthetic and real-world imaging artifacts, as well as segmentation of various pathological lesions across multiple MRI sequences, consistently demonstrate the effectiveness of our proposed method.
Continuous Online Adaptation Driven by User Interaction for Medical Image Segmentation
Mar 09, 2025Abstract:Interactive segmentation models use real-time user interactions, such as mouse clicks, as extra inputs to dynamically refine the model predictions. After model deployment, user corrections of model predictions could be used to adapt the model to the post-deployment data distribution, countering distribution-shift and enhancing reliability. Motivated by this, we introduce an online adaptation framework that enables an interactive segmentation model to continuously learn from user interaction and improve its performance on new data distributions, as it processes a sequence of test images. We introduce the Gaussian Point Loss function to train the model how to leverage user clicks, along with a two-stage online optimization method that adapts the model using the corrected predictions generated via user interactions. We demonstrate that this simple and therefore practical approach is very effective. Experiments on 5 fundus and 4 brain MRI databases demonstrate that our method outperforms existing approaches under various data distribution shifts, including segmentation of image modalities and pathologies not seen during training.
Feasibility of Federated Learning from Client Databases with Different Brain Diseases and MRI Modalities
Jun 17, 2024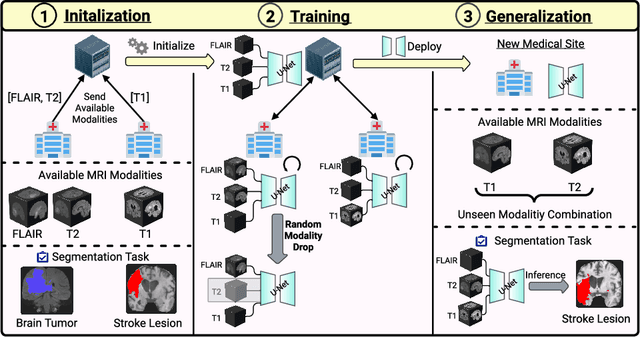
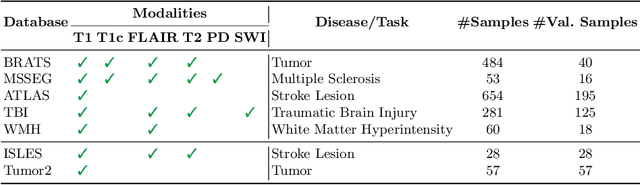
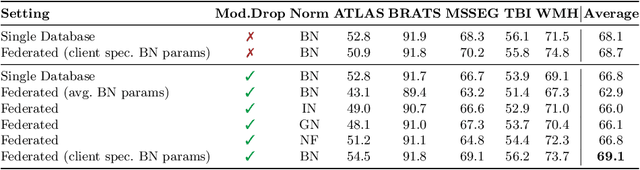
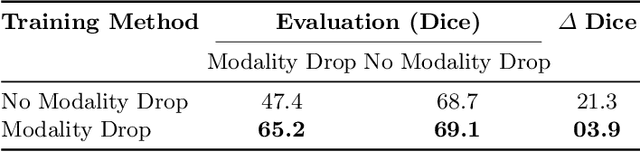
Abstract:Segmentation models for brain lesions in MRI are commonly developed for a specific disease and trained on data with a predefined set of MRI modalities. Each such model cannot segment the disease using data with a different set of MRI modalities, nor can it segment any other type of disease. Moreover, this training paradigm does not allow a model to benefit from learning from heterogeneous databases that may contain scans and segmentation labels for different types of brain pathologies and diverse sets of MRI modalities. Is it feasible to use Federated Learning (FL) for training a single model on client databases that contain scans and labels of different brain pathologies and diverse sets of MRI modalities? We demonstrate promising results by combining appropriate, simple, and practical modifications to the model and training strategy: Designing a model with input channels that cover the whole set of modalities available across clients, training with random modality drop, and exploring the effects of feature normalization methods. Evaluation on 7 brain MRI databases with 5 different diseases shows that such FL framework can train a single model that is shown to be very promising in segmenting all disease types seen during training. Importantly, it is able to segment these diseases in new databases that contain sets of modalities different from those in training clients. These results demonstrate, for the first time, feasibility and effectiveness of using FL to train a single segmentation model on decentralised data with diverse brain diseases and MRI modalities, a necessary step towards leveraging heterogeneous real-world databases. Code will be made available at: https://github.com/FelixWag/FL-MultiDisease-MRI
Feasibility and benefits of joint learning from MRI databases with different brain diseases and modalities for segmentation
May 28, 2024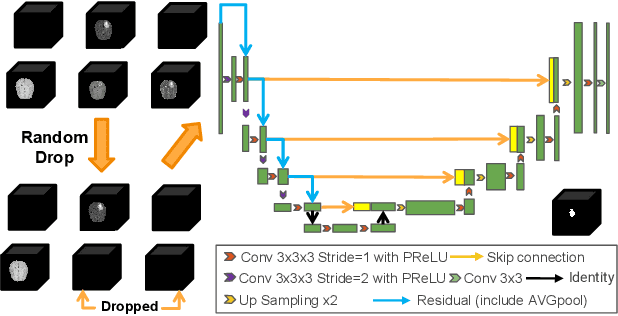
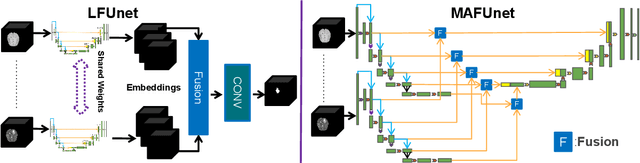

Abstract:Models for segmentation of brain lesions in multi-modal MRI are commonly trained for a specific pathology using a single database with a predefined set of MRI modalities, determined by a protocol for the specific disease. This work explores the following open questions: Is it feasible to train a model using multiple databases that contain varying sets of MRI modalities and annotations for different brain pathologies? Will this joint learning benefit performance on the sets of modalities and pathologies available during training? Will it enable analysis of new databases with different sets of modalities and pathologies? We develop and compare different methods and show that promising results can be achieved with appropriate, simple and practical alterations to the model and training framework. We experiment with 7 databases containing 5 types of brain pathologies and different sets of MRI modalities. Results demonstrate, for the first time, that joint training on multi-modal MRI databases with different brain pathologies and sets of modalities is feasible and offers practical benefits. It enables a single model to segment pathologies encountered during training in diverse sets of modalities, while facilitating segmentation of new types of pathologies such as via follow-up fine-tuning. The insights this study provides into the potential and limitations of this paradigm should prove useful for guiding future advances in the direction. Code and pretrained models: https://github.com/WenTXuL/MultiUnet
* Accepted to MIDL 2024
Revisiting Implicit Neural Representations in Low-Level Vision
Apr 20, 2023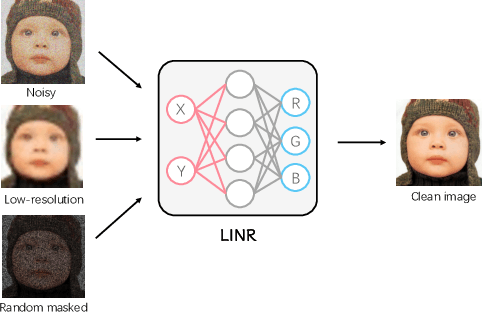
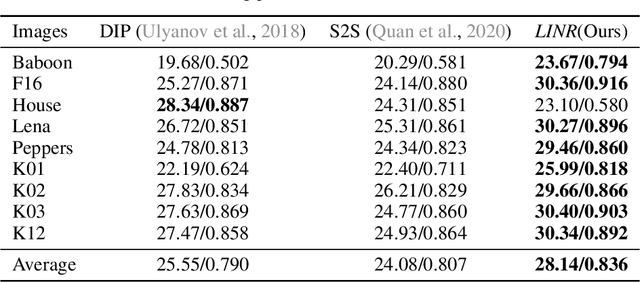
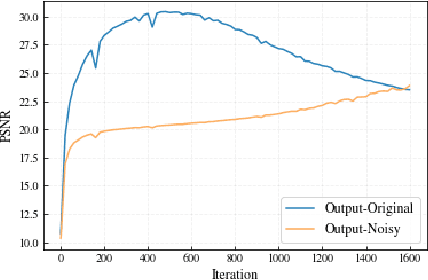
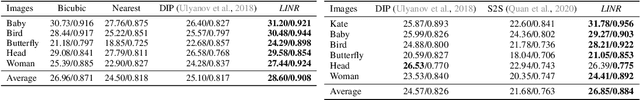
Abstract:Implicit Neural Representation (INR) has been emerging in computer vision in recent years. It has been shown to be effective in parameterising continuous signals such as dense 3D models from discrete image data, e.g. the neural radius field (NeRF). However, INR is under-explored in 2D image processing tasks. Considering the basic definition and the structure of INR, we are interested in its effectiveness in low-level vision problems such as image restoration. In this work, we revisit INR and investigate its application in low-level image restoration tasks including image denoising, super-resolution, inpainting, and deblurring. Extensive experimental evaluations suggest the superior performance of INR in several low-level vision tasks with limited resources, outperforming its counterparts by over 2dB. Code and models are available at https://github.com/WenTXuL/LINR
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge