Harry Anthony
Specialised or Generic? Tokenization Choices for Radiology Language Models
Aug 13, 2025Abstract:The vocabulary used by language models (LM) - defined by the tokenizer - plays a key role in text generation quality. However, its impact remains under-explored in radiology. In this work, we address this gap by systematically comparing general, medical, and domain-specific tokenizers on the task of radiology report summarisation across three imaging modalities. We also investigate scenarios with and without LM pre-training on PubMed abstracts. Our findings demonstrate that medical and domain-specific vocabularies outperformed widely used natural language alternatives when models are trained from scratch. Pre-training partially mitigates performance differences between tokenizers, whilst the domain-specific tokenizers achieve the most favourable results. Domain-specific tokenizers also reduce memory requirements due to smaller vocabularies and shorter sequences. These results demonstrate that adapting the vocabulary of LMs to the clinical domain provides practical benefits, including improved performance and reduced computational demands, making such models more accessible and effective for both research and real-world healthcare settings.
DIsoN: Decentralized Isolation Networks for Out-of-Distribution Detection in Medical Imaging
Jun 10, 2025



Abstract:Safe deployment of machine learning (ML) models in safety-critical domains such as medical imaging requires detecting inputs with characteristics not seen during training, known as out-of-distribution (OOD) detection, to prevent unreliable predictions. Effective OOD detection after deployment could benefit from access to the training data, enabling direct comparison between test samples and the training data distribution to identify differences. State-of-the-art OOD detection methods, however, either discard training data after deployment or assume that test samples and training data are centrally stored together, an assumption that rarely holds in real-world settings. This is because shipping training data with the deployed model is usually impossible due to the size of training databases, as well as proprietary or privacy constraints. We introduce the Isolation Network, an OOD detection framework that quantifies the difficulty of separating a target test sample from the training data by solving a binary classification task. We then propose Decentralized Isolation Networks (DIsoN), which enables the comparison of training and test data when data-sharing is impossible, by exchanging only model parameters between the remote computational nodes of training and deployment. We further extend DIsoN with class-conditioning, comparing a target sample solely with training data of its predicted class. We evaluate DIsoN on four medical imaging datasets (dermatology, chest X-ray, breast ultrasound, histopathology) across 12 OOD detection tasks. DIsoN performs favorably against existing methods while respecting data-privacy. This decentralized OOD detection framework opens the way for a new type of service that ML developers could provide along with their models: providing remote, secure utilization of their training data for OOD detection services. Code will be available upon acceptance at: *****
Continuous Online Adaptation Driven by User Interaction for Medical Image Segmentation
Mar 09, 2025Abstract:Interactive segmentation models use real-time user interactions, such as mouse clicks, as extra inputs to dynamically refine the model predictions. After model deployment, user corrections of model predictions could be used to adapt the model to the post-deployment data distribution, countering distribution-shift and enhancing reliability. Motivated by this, we introduce an online adaptation framework that enables an interactive segmentation model to continuously learn from user interaction and improve its performance on new data distributions, as it processes a sequence of test images. We introduce the Gaussian Point Loss function to train the model how to leverage user clicks, along with a two-stage online optimization method that adapts the model using the corrected predictions generated via user interactions. We demonstrate that this simple and therefore practical approach is very effective. Experiments on 5 fundus and 4 brain MRI databases demonstrate that our method outperforms existing approaches under various data distribution shifts, including segmentation of image modalities and pathologies not seen during training.
Evaluating Reliability in Medical DNNs: A Critical Analysis of Feature and Confidence-Based OOD Detection
Aug 30, 2024Abstract:Reliable use of deep neural networks (DNNs) for medical image analysis requires methods to identify inputs that differ significantly from the training data, called out-of-distribution (OOD), to prevent erroneous predictions. OOD detection methods can be categorised as either confidence-based (using the model's output layer for OOD detection) or feature-based (not using the output layer). We created two new OOD benchmarks by dividing the D7P (dermatology) and BreastMNIST (ultrasound) datasets into subsets which either contain or don't contain an artefact (rulers or annotations respectively). Models were trained with artefact-free images, and images with the artefacts were used as OOD test sets. For each OOD image, we created a counterfactual by manually removing the artefact via image processing, to assess the artefact's impact on the model's predictions. We show that OOD artefacts can boost a model's softmax confidence in its predictions, due to correlations in training data among other factors. This contradicts the common assumption that OOD artefacts should lead to more uncertain outputs, an assumption on which most confidence-based methods rely. We use this to explain why feature-based methods (e.g. Mahalanobis score) typically have greater OOD detection performance than confidence-based methods (e.g. MCP). However, we also show that feature-based methods typically perform worse at distinguishing between inputs that lead to correct and incorrect predictions (for both OOD and ID data). Following from these insights, we argue that a combination of feature-based and confidence-based methods should be used within DNN pipelines to mitigate their respective weaknesses. These project's code and OOD benchmarks are available at: https://github.com/HarryAnthony/Evaluating_OOD_detection.
On the use of Mahalanobis distance for out-of-distribution detection with neural networks for medical imaging
Sep 04, 2023Abstract:Implementing neural networks for clinical use in medical applications necessitates the ability for the network to detect when input data differs significantly from the training data, with the aim of preventing unreliable predictions. The community has developed several methods for out-of-distribution (OOD) detection, within which distance-based approaches - such as Mahalanobis distance - have shown potential. This paper challenges the prevailing community understanding that there is an optimal layer, or combination of layers, of a neural network for applying Mahalanobis distance for detection of any OOD pattern. Using synthetic artefacts to emulate OOD patterns, this paper shows the optimum layer to apply Mahalanobis distance changes with the type of OOD pattern, showing there is no one-fits-all solution. This paper also shows that separating this OOD detector into multiple detectors at different depths of the network can enhance the robustness for detecting different OOD patterns. These insights were validated on real-world OOD tasks, training models on CheXpert chest X-rays with no support devices, then using scans with unseen pacemakers (we manually labelled 50% of CheXpert for this research) and unseen sex as OOD cases. The results inform best-practices for the use of Mahalanobis distance for OOD detection. The manually annotated pacemaker labels and the project's code are available at: https://github.com/HarryAnthony/Mahalanobis-OOD-detection.
Modality Cycles with Masked Conditional Diffusion for Unsupervised Anomaly Segmentation in MRI
Aug 30, 2023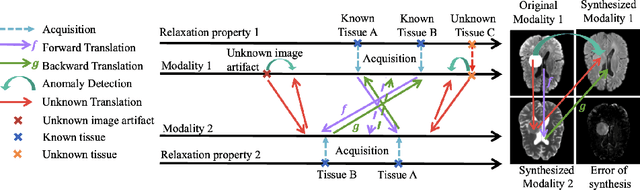
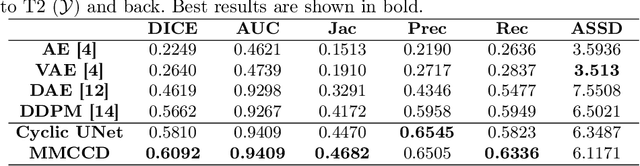
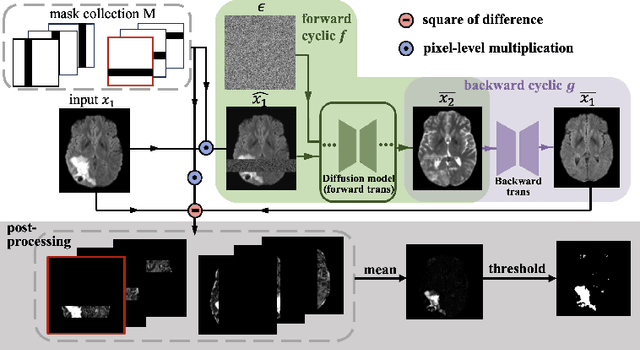
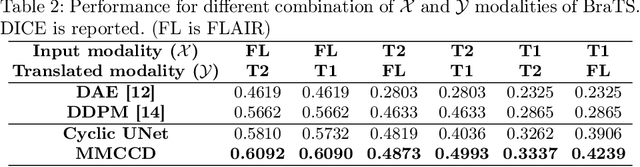
Abstract:Unsupervised anomaly segmentation aims to detect patterns that are distinct from any patterns processed during training, commonly called abnormal or out-of-distribution patterns, without providing any associated manual segmentations. Since anomalies during deployment can lead to model failure, detecting the anomaly can enhance the reliability of models, which is valuable in high-risk domains like medical imaging. This paper introduces Masked Modality Cycles with Conditional Diffusion (MMCCD), a method that enables segmentation of anomalies across diverse patterns in multimodal MRI. The method is based on two fundamental ideas. First, we propose the use of cyclic modality translation as a mechanism for enabling abnormality detection. Image-translation models learn tissue-specific modality mappings, which are characteristic of tissue physiology. Thus, these learned mappings fail to translate tissues or image patterns that have never been encountered during training, and the error enables their segmentation. Furthermore, we combine image translation with a masked conditional diffusion model, which attempts to `imagine' what tissue exists under a masked area, further exposing unknown patterns as the generative model fails to recreate them. We evaluate our method on a proxy task by training on healthy-looking slices of BraTS2021 multi-modality MRIs and testing on slices with tumors. We show that our method compares favorably to previous unsupervised approaches based on image reconstruction and denoising with autoencoders and diffusion models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge