Mingzhou Jiang
Topology-aware Pathological Consistency Matching for Weakly-Paired IHC Virtual Staining
Jan 06, 2026Abstract:Immunohistochemical (IHC) staining provides crucial molecular characterization of tissue samples and plays an indispensable role in the clinical examination and diagnosis of cancers. However, compared with the commonly used Hematoxylin and Eosin (H&E) staining, IHC staining involves complex procedures and is both time-consuming and expensive, which limits its widespread clinical use. Virtual staining converts H&E images to IHC images, offering a cost-effective alternative to clinical IHC staining. Nevertheless, using adjacent slides as ground truth often results in weakly-paired data with spatial misalignment and local deformations, hindering effective supervised learning. To address these challenges, we propose a novel topology-aware framework for H&E-to-IHC virtual staining. Specifically, we introduce a Topology-aware Consistency Matching (TACM) mechanism that employs graph contrastive learning and topological perturbations to learn robust matching patterns despite spatial misalignments, ensuring structural consistency. Furthermore, we propose a Topology-constrained Pathological Matching (TCPM) mechanism that aligns pathological positive regions based on node importance to enhance pathological consistency. Extensive experiments on two benchmarks across four staining tasks demonstrate that our method outperforms state-of-the-art approaches, achieving superior generation quality with higher clinical relevance.
MGI: Multimodal Contrastive pre-training of Genomic and Medical Imaging
Jun 02, 2024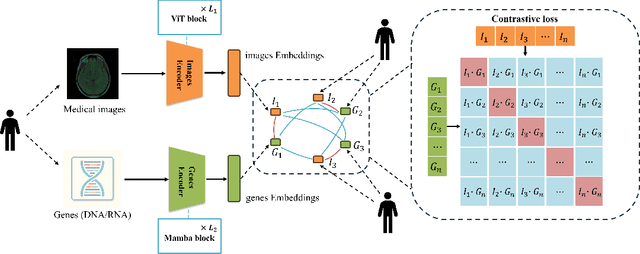
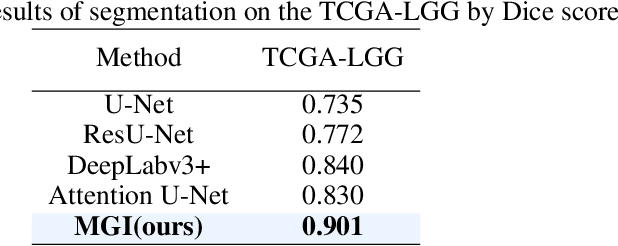
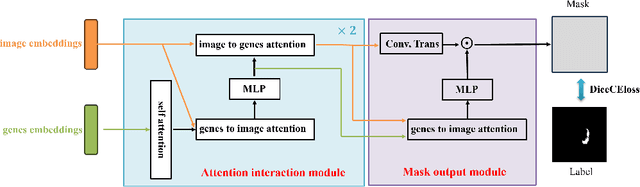
Abstract:Medicine is inherently a multimodal discipline. Medical images can reflect the pathological changes of cancer and tumors, while the expression of specific genes can influence their morphological characteristics. However, most deep learning models employed for these medical tasks are unimodal, making predictions using either image data or genomic data exclusively. In this paper, we propose a multimodal pre-training framework that jointly incorporates genomics and medical images for downstream tasks. To address the issues of high computational complexity and difficulty in capturing long-range dependencies in genes sequence modeling with MLP or Transformer architectures, we utilize Mamba to model these long genomic sequences. We aligns medical images and genes using a self-supervised contrastive learning approach which combines the Mamba as a genetic encoder and the Vision Transformer (ViT) as a medical image encoder. We pre-trained on the TCGA dataset using paired gene expression data and imaging data, and fine-tuned it for downstream tumor segmentation tasks. The results show that our model outperformed a wide range of related methods.
Uncertainty-Aware Adapter: Adapting Segment Anything Model (SAM) for Ambiguous Medical Image Segmentation
Mar 19, 2024



Abstract:The Segment Anything Model (SAM) gained significant success in natural image segmentation, and many methods have tried to fine-tune it to medical image segmentation. An efficient way to do so is by using Adapters, specialized modules that learn just a few parameters to tailor SAM specifically for medical images. However, unlike natural images, many tissues and lesions in medical images have blurry boundaries and may be ambiguous. Previous efforts to adapt SAM ignore this challenge and can only predict distinct segmentation. It may mislead clinicians or cause misdiagnosis, especially when encountering rare variants or situations with low model confidence. In this work, we propose a novel module called the Uncertainty-aware Adapter, which efficiently fine-tuning SAM for uncertainty-aware medical image segmentation. Utilizing a conditional variational autoencoder, we encoded stochastic samples to effectively represent the inherent uncertainty in medical imaging. We designed a new module on a standard adapter that utilizes a condition-based strategy to interact with samples to help SAM integrate uncertainty. We evaluated our method on two multi-annotated datasets with different modalities: LIDC-IDRI (lung abnormalities segmentation) and REFUGE2 (optic-cup segmentation). The experimental results show that the proposed model outperforms all the previous methods and achieves the new state-of-the-art (SOTA) on both benchmarks. We also demonstrated that our method can generate diverse segmentation hypotheses that are more realistic as well as heterogeneous.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge