Jun Wei
Testing the Fault-Tolerance of Multi-Sensor Fusion Perception in Autonomous Driving Systems
Apr 18, 2025Abstract:High-level Autonomous Driving Systems (ADSs), such as Google Waymo and Baidu Apollo, typically rely on multi-sensor fusion (MSF) based approaches to perceive their surroundings. This strategy increases perception robustness by combining the respective strengths of the camera and LiDAR and directly affects the safety-critical driving decisions of autonomous vehicles (AVs). However, in real-world autonomous driving scenarios, cameras and LiDAR are subject to various faults, which can probably significantly impact the decision-making and behaviors of ADSs. Existing MSF testing approaches only discovered corner cases that the MSF-based perception cannot accurately detected by MSF-based perception, while lacking research on how sensor faults affect the system-level behaviors of ADSs. To address this gap, we conduct the first exploration of the fault tolerance of MSF perception-based ADS for sensor faults. In this paper, we systematically and comprehensively build fault models for cameras and LiDAR in AVs and inject them into the MSF perception-based ADS to test its behaviors in test scenarios. To effectively and efficiently explore the parameter spaces of sensor fault models, we design a feedback-guided differential fuzzer to discover the safety violations of MSF perception-based ADS caused by the injected sensor faults. We evaluate FADE on the representative and practical industrial ADS, Baidu Apollo. Our evaluation results demonstrate the effectiveness and efficiency of FADE, and we conclude some useful findings from the experimental results. To validate the findings in the physical world, we use a real Baidu Apollo 6.0 EDU autonomous vehicle to conduct the physical experiments, and the results show the practical significance of our findings.
Simultaneous Polysomnography and Cardiotocography Reveal Temporal Correlation Between Maternal Obstructive Sleep Apnea and Fetal Hypoxia
Apr 17, 2025Abstract:Background: Obstructive sleep apnea syndrome (OSAS) during pregnancy is common and can negatively affect fetal outcomes. However, studies on the immediate effects of maternal hypoxia on fetal heart rate (FHR) changes are lacking. Methods: We used time-synchronized polysomnography (PSG) and cardiotocography (CTG) data from two cohorts to analyze the correlation between maternal hypoxia and FHR changes (accelerations or decelerations). Maternal hypoxic event characteristics were analyzed using generalized linear modeling (GLM) to assess their associations with different FHR changes. Results: A total of 118 pregnant women participated. FHR changes were significantly associated with maternal hypoxia, primarily characterized by accelerations. A longer hypoxic duration correlated with more significant FHR accelerations (P < 0.05), while prolonged hypoxia and greater SpO2 drop were linked to FHR decelerations (P < 0.05). Both cohorts showed a transient increase in FHR during maternal hypoxia, which returned to baseline after the event resolved. Conclusion: Maternal hypoxia significantly affects FHR, suggesting that maternal OSAS may contribute to fetal hypoxia. These findings highlight the importance of maternal-fetal interactions and provide insights for future interventions.
MARIO: A Mixed Annotation Framework For Polyp Segmentation
Jan 19, 2025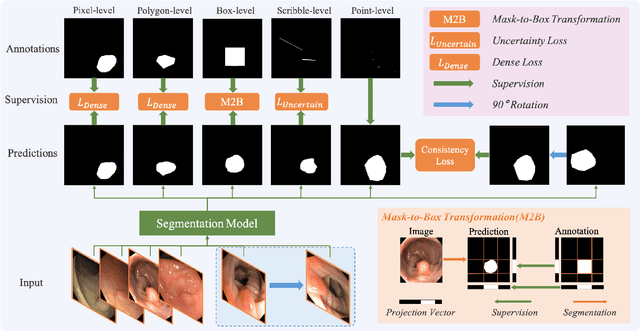
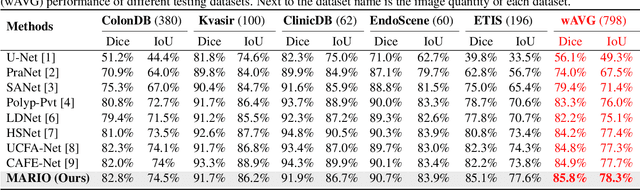
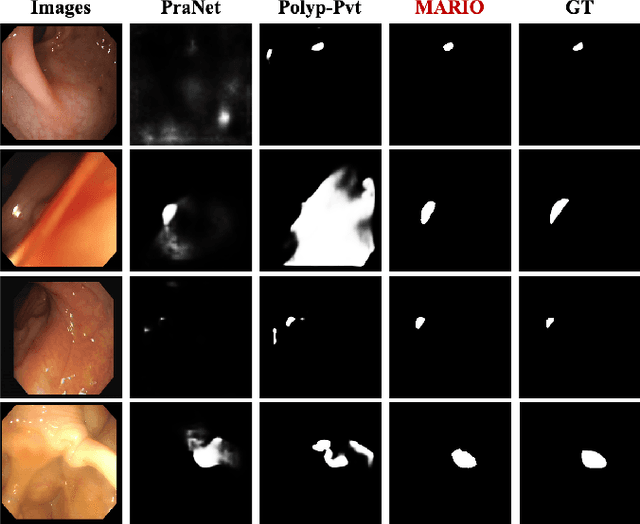

Abstract:Existing polyp segmentation models are limited by high labeling costs and the small size of datasets. Additionally, vast polyp datasets remain underutilized because these models typically rely on a single type of annotation. To address this dilemma, we introduce MARIO, a mixed supervision model designed to accommodate various annotation types, significantly expanding the range of usable data. MARIO learns from underutilized datasets by incorporating five forms of supervision: pixel-level, box-level, polygon-level, scribblelevel, and point-level. Each form of supervision is associated with a tailored loss that effectively leverages the supervision labels while minimizing the noise. This allows MARIO to move beyond the constraints of relying on a single annotation type. Furthermore, MARIO primarily utilizes dataset with weak and cheap annotations, reducing the dependence on large-scale, fully annotated ones. Experimental results across five benchmark datasets demonstrate that MARIO consistently outperforms existing methods, highlighting its efficacy in balancing trade-offs between different forms of supervision and maximizing polyp segmentation performance
An LLM-Empowered Adaptive Evolutionary Algorithm For Multi-Component Deep Learning Systems
Jan 01, 2025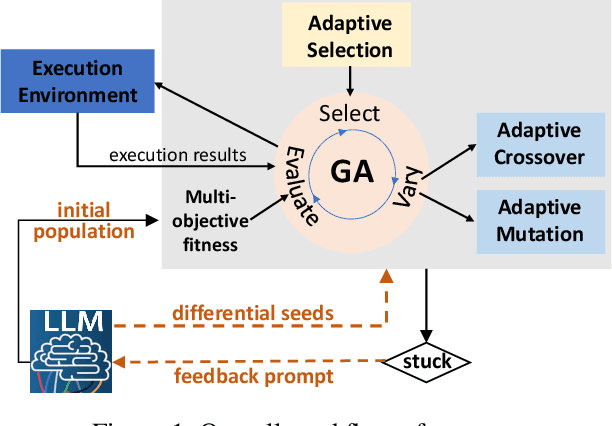


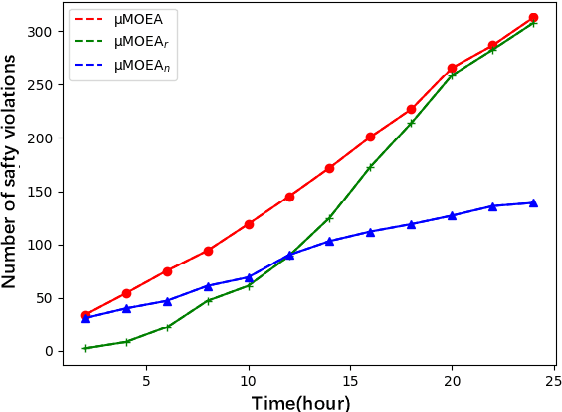
Abstract:Multi-objective evolutionary algorithms (MOEAs) are widely used for searching optimal solutions in complex multi-component applications. Traditional MOEAs for multi-component deep learning (MCDL) systems face challenges in enhancing the search efficiency while maintaining the diversity. To combat these, this paper proposes $\mu$MOEA, the first LLM-empowered adaptive evolutionary search algorithm to detect safety violations in MCDL systems. Inspired by the context-understanding ability of Large Language Models (LLMs), $\mu$MOEA promotes the LLM to comprehend the optimization problem and generate an initial population tailed to evolutionary objectives. Subsequently, it employs adaptive selection and variation to iteratively produce offspring, balancing the evolutionary efficiency and diversity. During the evolutionary process, to navigate away from the local optima, $\mu$MOEA integrates the evolutionary experience back into the LLM. This utilization harnesses the LLM's quantitative reasoning prowess to generate differential seeds, breaking away from current optimal solutions. We evaluate $\mu$MOEA in finding safety violations of MCDL systems, and compare its performance with state-of-the-art MOEA methods. Experimental results show that $\mu$MOEA can significantly improve the efficiency and diversity of the evolutionary search.
Generative Semantic Communication for Joint Image Transmission and Segmentation
Nov 27, 2024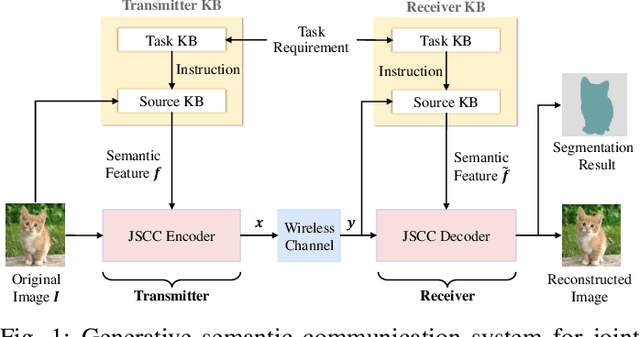
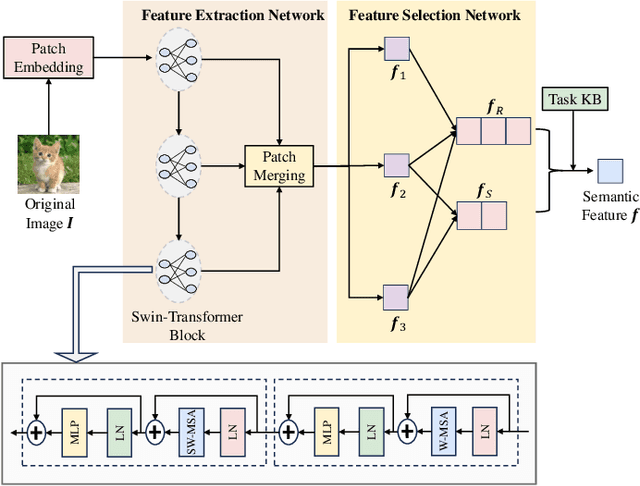
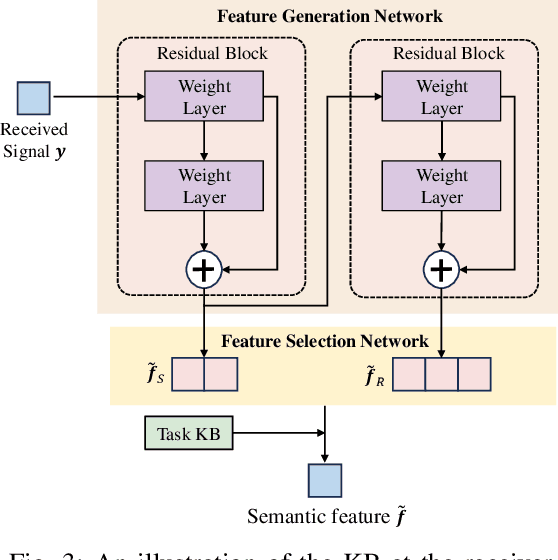
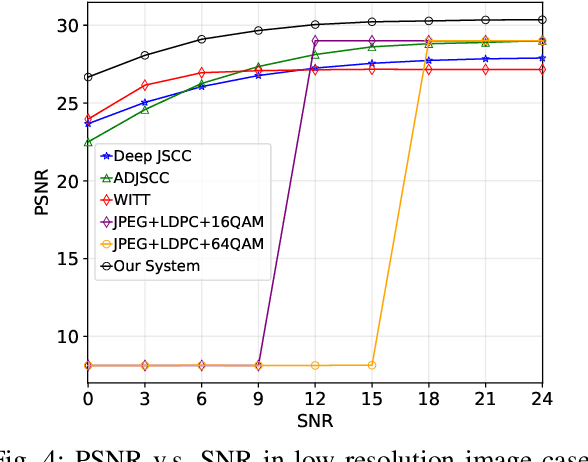
Abstract:Semantic communication has emerged as a promising technology for enhancing communication efficiency. However, most existing research emphasizes single-task reconstruction, neglecting model adaptability and generalization across multi-task systems. In this paper, we propose a novel generative semantic communication system that supports both image reconstruction and segmentation tasks. Our approach builds upon semantic knowledge bases (KBs) at both the transmitter and receiver, with each semantic KB comprising a source KB and a task KB. The source KB at the transmitter leverages a hierarchical Swin-Transformer, a generative AI scheme, to extract multi-level features from the input image. Concurrently, the counterpart source KB at the receiver utilizes hierarchical residual blocks to generate task-specific knowledge. Furthermore, the two task KBs adopt a semantic similarity model to map different task requirements into pre-defined task instructions, thereby facilitating the feature selection of the source KBs. Additionally, we develop a unified residual block-based joint source and channel (JSCC) encoder and two task-specific JSCC decoders to achieve the two image tasks. In particular, a generative diffusion model is adopted to construct the JSCC decoder for the image reconstruction task. Experimental results demonstrate that our multi-task generative semantic communication system outperforms previous single-task communication systems in terms of peak signal-to-noise ratio and segmentation accuracy.
Privacy-Preserving Federated Foundation Model for Generalist Ultrasound Artificial Intelligence
Nov 25, 2024



Abstract:Ultrasound imaging is widely used in clinical diagnosis due to its non-invasive nature and real-time capabilities. However, conventional ultrasound diagnostics face several limitations, including high dependence on physician expertise and suboptimal image quality, which complicates interpretation and increases the likelihood of diagnostic errors. Artificial intelligence (AI) has emerged as a promising solution to enhance clinical diagnosis, particularly in detecting abnormalities across various biomedical imaging modalities. Nonetheless, current AI models for ultrasound imaging face critical challenges. First, these models often require large volumes of labeled medical data, raising concerns over patient privacy breaches. Second, most existing models are task-specific, which restricts their broader clinical utility. To overcome these challenges, we present UltraFedFM, an innovative privacy-preserving ultrasound foundation model. UltraFedFM is collaboratively pre-trained using federated learning across 16 distributed medical institutions in 9 countries, leveraging a dataset of over 1 million ultrasound images covering 19 organs and 10 ultrasound modalities. This extensive and diverse data, combined with a secure training framework, enables UltraFedFM to exhibit strong generalization and diagnostic capabilities. It achieves an average area under the receiver operating characteristic curve of 0.927 for disease diagnosis and a dice similarity coefficient of 0.878 for lesion segmentation. Notably, UltraFedFM surpasses the diagnostic accuracy of mid-level ultrasonographers and matches the performance of expert-level sonographers in the joint diagnosis of 8 common systemic diseases. These findings indicate that UltraFedFM can significantly enhance clinical diagnostics while safeguarding patient privacy, marking an advancement in AI-driven ultrasound imaging for future clinical applications.
MixPolyp: Integrating Mask, Box and Scribble Supervision for Enhanced Polyp Segmentation
Sep 25, 2024



Abstract:Limited by the expensive labeling, polyp segmentation models are plagued by data shortages. To tackle this, we propose the mixed supervised polyp segmentation paradigm (MixPolyp). Unlike traditional models relying on a single type of annotation, MixPolyp combines diverse annotation types (mask, box, and scribble) within a single model, thereby expanding the range of available data and reducing labeling costs. To achieve this, MixPolyp introduces three novel supervision losses to handle various annotations: Subspace Projection loss (L_SP), Binary Minimum Entropy loss (L_BME), and Linear Regularization loss (L_LR). For box annotations, L_SP eliminates shape inconsistencies between the prediction and the supervision. For scribble annotations, L_BME provides supervision for unlabeled pixels through minimum entropy constraint, thereby alleviating supervision sparsity. Furthermore, L_LR provides dense supervision by enforcing consistency among the predictions, thus reducing the non-uniqueness. These losses are independent of the model structure, making them generally applicable. They are used only during training, adding no computational cost during inference. Extensive experiments on five datasets demonstrate MixPolyp's effectiveness.
Let Video Teaches You More: Video-to-Image Knowledge Distillation using DEtection TRansformer for Medical Video Lesion Detection
Aug 26, 2024



Abstract:AI-assisted lesion detection models play a crucial role in the early screening of cancer. However, previous image-based models ignore the inter-frame contextual information present in videos. On the other hand, video-based models capture the inter-frame context but are computationally expensive. To mitigate this contradiction, we delve into Video-to-Image knowledge distillation leveraging DEtection TRansformer (V2I-DETR) for the task of medical video lesion detection. V2I-DETR adopts a teacher-student network paradigm. The teacher network aims at extracting temporal contexts from multiple frames and transferring them to the student network, and the student network is an image-based model dedicated to fast prediction in inference. By distilling multi-frame contexts into a single frame, the proposed V2I-DETR combines the advantages of utilizing temporal contexts from video-based models and the inference speed of image-based models. Through extensive experiments, V2I-DETR outperforms previous state-of-the-art methods by a large margin while achieving the real-time inference speed (30 FPS) as the image-based model.
Towards a Benchmark for Colorectal Cancer Segmentation in Endorectal Ultrasound Videos: Dataset and Model Development
Aug 19, 2024Abstract:Endorectal ultrasound (ERUS) is an important imaging modality that provides high reliability for diagnosing the depth and boundary of invasion in colorectal cancer. However, the lack of a large-scale ERUS dataset with high-quality annotations hinders the development of automatic ultrasound diagnostics. In this paper, we collected and annotated the first benchmark dataset that covers diverse ERUS scenarios, i.e. colorectal cancer segmentation, detection, and infiltration depth staging. Our ERUS-10K dataset comprises 77 videos and 10,000 high-resolution annotated frames. Based on this dataset, we further introduce a benchmark model for colorectal cancer segmentation, named the Adaptive Sparse-context TRansformer (ASTR). ASTR is designed based on three considerations: scanning mode discrepancy, temporal information, and low computational complexity. For generalizing to different scanning modes, the adaptive scanning-mode augmentation is proposed to convert between raw sector images and linear scan ones. For mining temporal information, the sparse-context transformer is incorporated to integrate inter-frame local and global features. For reducing computational complexity, the sparse-context block is introduced to extract contextual features from auxiliary frames. Finally, on the benchmark dataset, the proposed ASTR model achieves a 77.6% Dice score in rectal cancer segmentation, largely outperforming previous state-of-the-art methods.
ScribblePolyp: Scribble-Supervised Polyp Segmentation through Dual Consistency Alignment
Nov 09, 2023



Abstract:Automatic polyp segmentation models play a pivotal role in the clinical diagnosis of gastrointestinal diseases. In previous studies, most methods relied on fully supervised approaches, necessitating pixel-level annotations for model training. However, the creation of pixel-level annotations is both expensive and time-consuming, impeding the development of model generalization. In response to this challenge, we introduce ScribblePolyp, a novel scribble-supervised polyp segmentation framework. Unlike fully-supervised models, ScribblePolyp only requires the annotation of two lines (scribble labels) for each image, significantly reducing the labeling cost. Despite the coarse nature of scribble labels, which leave a substantial portion of pixels unlabeled, we propose a two-branch consistency alignment approach to provide supervision for these unlabeled pixels. The first branch employs transformation consistency alignment to narrow the gap between predictions under different transformations of the same input image. The second branch leverages affinity propagation to refine predictions into a soft version, extending additional supervision to unlabeled pixels. In summary, ScribblePolyp is an efficient model that does not rely on teacher models or moving average pseudo labels during training. Extensive experiments on the SUN-SEG dataset underscore the effectiveness of ScribblePolyp, achieving a Dice score of 0.8155, with the potential for a 1.8% improvement in the Dice score through a straightforward self-training strategy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge