Zixun Zhang
Privacy-Preserving Federated Foundation Model for Generalist Ultrasound Artificial Intelligence
Nov 25, 2024



Abstract:Ultrasound imaging is widely used in clinical diagnosis due to its non-invasive nature and real-time capabilities. However, conventional ultrasound diagnostics face several limitations, including high dependence on physician expertise and suboptimal image quality, which complicates interpretation and increases the likelihood of diagnostic errors. Artificial intelligence (AI) has emerged as a promising solution to enhance clinical diagnosis, particularly in detecting abnormalities across various biomedical imaging modalities. Nonetheless, current AI models for ultrasound imaging face critical challenges. First, these models often require large volumes of labeled medical data, raising concerns over patient privacy breaches. Second, most existing models are task-specific, which restricts their broader clinical utility. To overcome these challenges, we present UltraFedFM, an innovative privacy-preserving ultrasound foundation model. UltraFedFM is collaboratively pre-trained using federated learning across 16 distributed medical institutions in 9 countries, leveraging a dataset of over 1 million ultrasound images covering 19 organs and 10 ultrasound modalities. This extensive and diverse data, combined with a secure training framework, enables UltraFedFM to exhibit strong generalization and diagnostic capabilities. It achieves an average area under the receiver operating characteristic curve of 0.927 for disease diagnosis and a dice similarity coefficient of 0.878 for lesion segmentation. Notably, UltraFedFM surpasses the diagnostic accuracy of mid-level ultrasonographers and matches the performance of expert-level sonographers in the joint diagnosis of 8 common systemic diseases. These findings indicate that UltraFedFM can significantly enhance clinical diagnostics while safeguarding patient privacy, marking an advancement in AI-driven ultrasound imaging for future clinical applications.
Let Video Teaches You More: Video-to-Image Knowledge Distillation using DEtection TRansformer for Medical Video Lesion Detection
Aug 26, 2024



Abstract:AI-assisted lesion detection models play a crucial role in the early screening of cancer. However, previous image-based models ignore the inter-frame contextual information present in videos. On the other hand, video-based models capture the inter-frame context but are computationally expensive. To mitigate this contradiction, we delve into Video-to-Image knowledge distillation leveraging DEtection TRansformer (V2I-DETR) for the task of medical video lesion detection. V2I-DETR adopts a teacher-student network paradigm. The teacher network aims at extracting temporal contexts from multiple frames and transferring them to the student network, and the student network is an image-based model dedicated to fast prediction in inference. By distilling multi-frame contexts into a single frame, the proposed V2I-DETR combines the advantages of utilizing temporal contexts from video-based models and the inference speed of image-based models. Through extensive experiments, V2I-DETR outperforms previous state-of-the-art methods by a large margin while achieving the real-time inference speed (30 FPS) as the image-based model.
Towards a Benchmark for Colorectal Cancer Segmentation in Endorectal Ultrasound Videos: Dataset and Model Development
Aug 19, 2024Abstract:Endorectal ultrasound (ERUS) is an important imaging modality that provides high reliability for diagnosing the depth and boundary of invasion in colorectal cancer. However, the lack of a large-scale ERUS dataset with high-quality annotations hinders the development of automatic ultrasound diagnostics. In this paper, we collected and annotated the first benchmark dataset that covers diverse ERUS scenarios, i.e. colorectal cancer segmentation, detection, and infiltration depth staging. Our ERUS-10K dataset comprises 77 videos and 10,000 high-resolution annotated frames. Based on this dataset, we further introduce a benchmark model for colorectal cancer segmentation, named the Adaptive Sparse-context TRansformer (ASTR). ASTR is designed based on three considerations: scanning mode discrepancy, temporal information, and low computational complexity. For generalizing to different scanning modes, the adaptive scanning-mode augmentation is proposed to convert between raw sector images and linear scan ones. For mining temporal information, the sparse-context transformer is incorporated to integrate inter-frame local and global features. For reducing computational complexity, the sparse-context block is introduced to extract contextual features from auxiliary frames. Finally, on the benchmark dataset, the proposed ASTR model achieves a 77.6% Dice score in rectal cancer segmentation, largely outperforming previous state-of-the-art methods.
Joint Signal Detection and Automatic Modulation Classification via Deep Learning
Apr 29, 2024



Abstract:Signal detection and modulation classification are two crucial tasks in various wireless communication systems. Different from prior works that investigate them independently, this paper studies the joint signal detection and automatic modulation classification (AMC) by considering a realistic and complex scenario, in which multiple signals with different modulation schemes coexist at different carrier frequencies. We first generate a coexisting RADIOML dataset (CRML23) to facilitate the joint design. Different from the publicly available AMC dataset ignoring the signal detection step and containing only one signal, our synthetic dataset covers the more realistic multiple-signal coexisting scenario. Then, we present a joint framework for detection and classification (JDM) for such a multiple-signal coexisting environment, which consists of two modules for signal detection and AMC, respectively. In particular, these two modules are interconnected using a designated data structure called "proposal". Finally, we conduct extensive simulations over the newly developed dataset, which demonstrate the effectiveness of our designs. Our code and dataset are now available as open-source (https://github.com/Singingkettle/ChangShuoRadioData).
ECC-PolypDet: Enhanced CenterNet with Contrastive Learning for Automatic Polyp Detection
Jan 10, 2024



Abstract:Accurate polyp detection is critical for early colorectal cancer diagnosis. Although remarkable progress has been achieved in recent years, the complex colon environment and concealed polyps with unclear boundaries still pose severe challenges in this area. Existing methods either involve computationally expensive context aggregation or lack prior modeling of polyps, resulting in poor performance in challenging cases. In this paper, we propose the Enhanced CenterNet with Contrastive Learning (ECC-PolypDet), a two-stage training \& end-to-end inference framework that leverages images and bounding box annotations to train a general model and fine-tune it based on the inference score to obtain a final robust model. Specifically, we conduct Box-assisted Contrastive Learning (BCL) during training to minimize the intra-class difference and maximize the inter-class difference between foreground polyps and backgrounds, enabling our model to capture concealed polyps. Moreover, to enhance the recognition of small polyps, we design the Semantic Flow-guided Feature Pyramid Network (SFFPN) to aggregate multi-scale features and the Heatmap Propagation (HP) module to boost the model's attention on polyp targets. In the fine-tuning stage, we introduce the IoU-guided Sample Re-weighting (ISR) mechanism to prioritize hard samples by adaptively adjusting the loss weight for each sample during fine-tuning. Extensive experiments on six large-scale colonoscopy datasets demonstrate the superiority of our model compared with previous state-of-the-art detectors.
ScribblePolyp: Scribble-Supervised Polyp Segmentation through Dual Consistency Alignment
Nov 09, 2023



Abstract:Automatic polyp segmentation models play a pivotal role in the clinical diagnosis of gastrointestinal diseases. In previous studies, most methods relied on fully supervised approaches, necessitating pixel-level annotations for model training. However, the creation of pixel-level annotations is both expensive and time-consuming, impeding the development of model generalization. In response to this challenge, we introduce ScribblePolyp, a novel scribble-supervised polyp segmentation framework. Unlike fully-supervised models, ScribblePolyp only requires the annotation of two lines (scribble labels) for each image, significantly reducing the labeling cost. Despite the coarse nature of scribble labels, which leave a substantial portion of pixels unlabeled, we propose a two-branch consistency alignment approach to provide supervision for these unlabeled pixels. The first branch employs transformation consistency alignment to narrow the gap between predictions under different transformations of the same input image. The second branch leverages affinity propagation to refine predictions into a soft version, extending additional supervision to unlabeled pixels. In summary, ScribblePolyp is an efficient model that does not rely on teacher models or moving average pseudo labels during training. Extensive experiments on the SUN-SEG dataset underscore the effectiveness of ScribblePolyp, achieving a Dice score of 0.8155, with the potential for a 1.8% improvement in the Dice score through a straightforward self-training strategy.
YONA: You Only Need One Adjacent Reference-frame for Accurate and Fast Video Polyp Detection
Jun 06, 2023



Abstract:Accurate polyp detection is essential for assisting clinical rectal cancer diagnoses. Colonoscopy videos contain richer information than still images, making them a valuable resource for deep learning methods. Great efforts have been made to conduct video polyp detection through multi-frame temporal/spatial aggregation. However, unlike common fixed-camera video, the camera-moving scene in colonoscopy videos can cause rapid video jitters, leading to unstable training for existing video detection models. Additionally, the concealed nature of some polyps and the complex background environment further hinder the performance of existing video detectors. In this paper, we propose the \textbf{YONA} (\textbf{Y}ou \textbf{O}nly \textbf{N}eed one \textbf{A}djacent Reference-frame) method, an efficient end-to-end training framework for video polyp detection. YONA fully exploits the information of one previous adjacent frame and conducts polyp detection on the current frame without multi-frame collaborations. Specifically, for the foreground, YONA adaptively aligns the current frame's channel activation patterns with its adjacent reference frames according to their foreground similarity. For the background, YONA conducts background dynamic alignment guided by inter-frame difference to eliminate the invalid features produced by drastic spatial jitters. Moreover, YONA applies cross-frame contrastive learning during training, leveraging the ground truth bounding box to improve the model's perception of polyp and background. Quantitative and qualitative experiments on three public challenging benchmarks demonstrate that our proposed YONA outperforms previous state-of-the-art competitors by a large margin in both accuracy and speed.
Hierarchical Weight Averaging for Deep Neural Networks
Apr 23, 2023



Abstract:Despite the simplicity, stochastic gradient descent (SGD)-like algorithms are successful in training deep neural networks (DNNs). Among various attempts to improve SGD, weight averaging (WA), which averages the weights of multiple models, has recently received much attention in the literature. Broadly, WA falls into two categories: 1) online WA, which averages the weights of multiple models trained in parallel, is designed for reducing the gradient communication overhead of parallel mini-batch SGD, and 2) offline WA, which averages the weights of one model at different checkpoints, is typically used to improve the generalization ability of DNNs. Though online and offline WA are similar in form, they are seldom associated with each other. Besides, these methods typically perform either offline parameter averaging or online parameter averaging, but not both. In this work, we firstly attempt to incorporate online and offline WA into a general training framework termed Hierarchical Weight Averaging (HWA). By leveraging both the online and offline averaging manners, HWA is able to achieve both faster convergence speed and superior generalization performance without any fancy learning rate adjustment. Besides, we also analyze the issues faced by existing WA methods, and how our HWA address them, empirically. Finally, extensive experiments verify that HWA outperforms the state-of-the-art methods significantly.
APAUNet: Axis Projection Attention UNet for Small Target in 3D Medical Segmentation
Oct 04, 2022

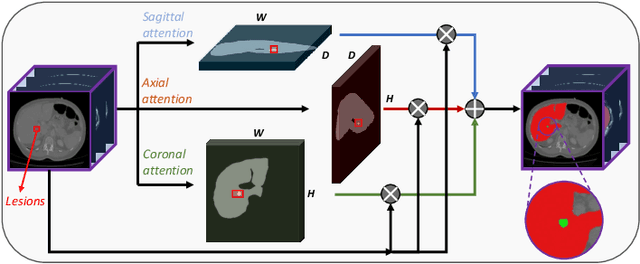

Abstract:In 3D medical image segmentation, small targets segmentation is crucial for diagnosis but still faces challenges. In this paper, we propose the Axis Projection Attention UNet, named APAUNet, for 3D medical image segmentation, especially for small targets. Considering the large proportion of the background in the 3D feature space, we introduce a projection strategy to project the 3D features into three orthogonal 2D planes to capture the contextual attention from different views. In this way, we can filter out the redundant feature information and mitigate the loss of critical information for small lesions in 3D scans. Then we utilize a dimension hybridization strategy to fuse the 3D features with attention from different axes and merge them by a weighted summation to adaptively learn the importance of different perspectives. Finally, in the APA Decoder, we concatenate both high and low resolution features in the 2D projection process, thereby obtaining more precise multi-scale information, which is vital for small lesion segmentation. Quantitative and qualitative experimental results on two public datasets (BTCV and MSD) demonstrate that our proposed APAUNet outperforms the other methods. Concretely, our APAUNet achieves an average dice score of 87.84 on BTCV, 84.48 on MSD-Liver and 69.13 on MSD-Pancreas, and significantly surpass the previous SOTA methods on small targets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge