Yuncheng Jiang
GRCF: Two-Stage Groupwise Ranking and Calibration Framework for Multimodal Sentiment Analysis
Jan 14, 2026Abstract:Most Multimodal Sentiment Analysis research has focused on point-wise regression. While straightforward, this approach is sensitive to label noise and neglects whether one sample is more positive than another, resulting in unstable predictions and poor correlation alignment. Pairwise ordinal learning frameworks emerged to address this gap, capturing relative order by learning from comparisons. Yet, they introduce two new trade-offs: First, they assign uniform importance to all comparisons, failing to adaptively focus on hard-to-rank samples. Second, they employ static ranking margins, which fail to reflect the varying semantic distances between sentiment groups. To address this, we propose a Two-Stage Group-wise Ranking and Calibration Framework (GRCF) that adapts the philosophy of Group Relative Policy Optimization (GRPO). Our framework resolves these trade-offs by simultaneously preserving relative ordinal structure, ensuring absolute score calibration, and adaptively focusing on difficult samples. Specifically, Stage 1 introduces a GRPO-inspired Advantage-Weighted Dynamic Margin Ranking Loss to build a fine-grained ordinal structure. Stage 2 then employs an MAE-driven objective to align prediction magnitudes. To validate its generalizability, we extend GRCF to classification tasks, including multimodal humor detection and sarcasm detection. GRCF achieves state-of-the-art performance on core regression benchmarks, while also showing strong generalizability in classification tasks.
Cross Modal Fine-Grained Alignment via Granularity-Aware and Region-Uncertain Modeling
Nov 19, 2025Abstract:Fine-grained image-text alignment is a pivotal challenge in multimodal learning, underpinning key applications such as visual question answering, image captioning, and vision-language navigation. Unlike global alignment, fine-grained alignment requires precise correspondence between localized visual regions and textual tokens, often hindered by noisy attention mechanisms and oversimplified modeling of cross-modal relationships. In this work, we identify two fundamental limitations of existing approaches: the lack of robust intra-modal mechanisms to assess the significance of visual and textual tokens, leading to poor generalization in complex scenes; and the absence of fine-grained uncertainty modeling, which fails to capture the one-to-many and many-to-one nature of region-word correspondences. To address these issues, we propose a unified approach that incorporates significance-aware and granularity-aware modeling and region-level uncertainty modeling. Our method leverages modality-specific biases to identify salient features without relying on brittle cross-modal attention, and represents region features as a mixture of Gaussian distributions to capture fine-grained uncertainty. Extensive experiments on Flickr30K and MS-COCO demonstrate that our approach achieves state-of-the-art performance across various backbone architectures, significantly enhancing the robustness and interpretability of fine-grained image-text alignment.
Beyond Cosine Similarity Magnitude-Aware CLIP for No-Reference Image Quality Assessment
Nov 13, 2025Abstract:Recent efforts have repurposed the Contrastive Language-Image Pre-training (CLIP) model for No-Reference Image Quality Assessment (NR-IQA) by measuring the cosine similarity between the image embedding and textual prompts such as "a good photo" or "a bad photo." However, this semantic similarity overlooks a critical yet underexplored cue: the magnitude of the CLIP image features, which we empirically find to exhibit a strong correlation with perceptual quality. In this work, we introduce a novel adaptive fusion framework that complements cosine similarity with a magnitude-aware quality cue. Specifically, we first extract the absolute CLIP image features and apply a Box-Cox transformation to statistically normalize the feature distribution and mitigate semantic sensitivity. The resulting scalar summary serves as a semantically-normalized auxiliary cue that complements cosine-based prompt matching. To integrate both cues effectively, we further design a confidence-guided fusion scheme that adaptively weighs each term according to its relative strength. Extensive experiments on multiple benchmark IQA datasets demonstrate that our method consistently outperforms standard CLIP-based IQA and state-of-the-art baselines, without any task-specific training.
Disentangling Bias by Modeling Intra- and Inter-modal Causal Attention for Multimodal Sentiment Analysis
Aug 07, 2025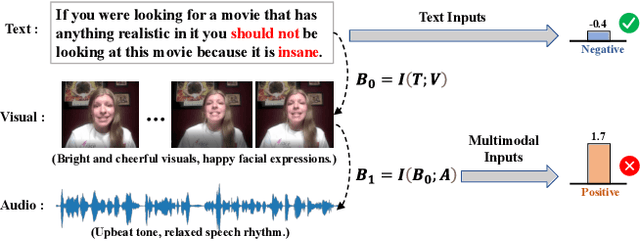
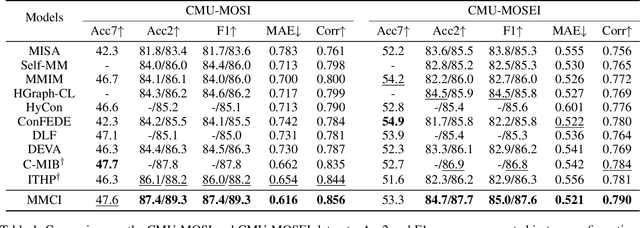
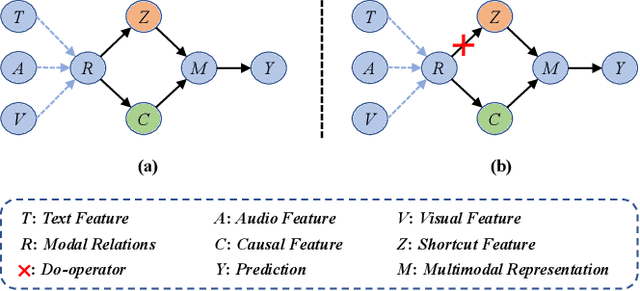
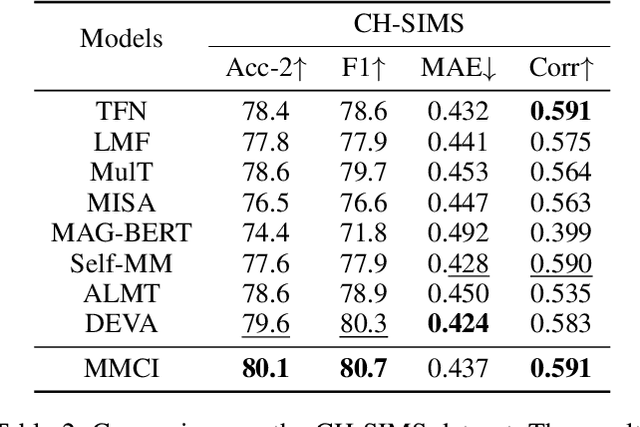
Abstract:Multimodal sentiment analysis (MSA) aims to understand human emotions by integrating information from multiple modalities, such as text, audio, and visual data. However, existing methods often suffer from spurious correlations both within and across modalities, leading models to rely on statistical shortcuts rather than true causal relationships, thereby undermining generalization. To mitigate this issue, we propose a Multi-relational Multimodal Causal Intervention (MMCI) model, which leverages the backdoor adjustment from causal theory to address the confounding effects of such shortcuts. Specifically, we first model the multimodal inputs as a multi-relational graph to explicitly capture intra- and inter-modal dependencies. Then, we apply an attention mechanism to separately estimate and disentangle the causal features and shortcut features corresponding to these intra- and inter-modal relations. Finally, by applying the backdoor adjustment, we stratify the shortcut features and dynamically combine them with the causal features to encourage MMCI to produce stable predictions under distribution shifts. Extensive experiments on several standard MSA datasets and out-of-distribution (OOD) test sets demonstrate that our method effectively suppresses biases and improves performance.
Towards Explainable Fusion and Balanced Learning in Multimodal Sentiment Analysis
Apr 16, 2025Abstract:Multimodal Sentiment Analysis (MSA) faces two critical challenges: the lack of interpretability in the decision logic of multimodal fusion and modality imbalance caused by disparities in inter-modal information density. To address these issues, we propose KAN-MCP, a novel framework that integrates the interpretability of Kolmogorov-Arnold Networks (KAN) with the robustness of the Multimodal Clean Pareto (MCPareto) framework. First, KAN leverages its univariate function decomposition to achieve transparent analysis of cross-modal interactions. This structural design allows direct inspection of feature transformations without relying on external interpretation tools, thereby ensuring both high expressiveness and interpretability. Second, the proposed MCPareto enhances robustness by addressing modality imbalance and noise interference. Specifically, we introduce the Dimensionality Reduction and Denoising Modal Information Bottleneck (DRD-MIB) method, which jointly denoises and reduces feature dimensionality. This approach provides KAN with discriminative low-dimensional inputs to reduce the modeling complexity of KAN while preserving critical sentiment-related information. Furthermore, MCPareto dynamically balances gradient contributions across modalities using the purified features output by DRD-MIB, ensuring lossless transmission of auxiliary signals and effectively alleviating modality imbalance. This synergy of interpretability and robustness not only achieves superior performance on benchmark datasets such as CMU-MOSI, CMU-MOSEI, and CH-SIMS v2 but also offers an intuitive visualization interface through KAN's interpretable architecture.
A Performance Investigation of Multimodal Multiobjective Optimization Algorithms in Solving Two Types of Real-World Problems
Dec 04, 2024

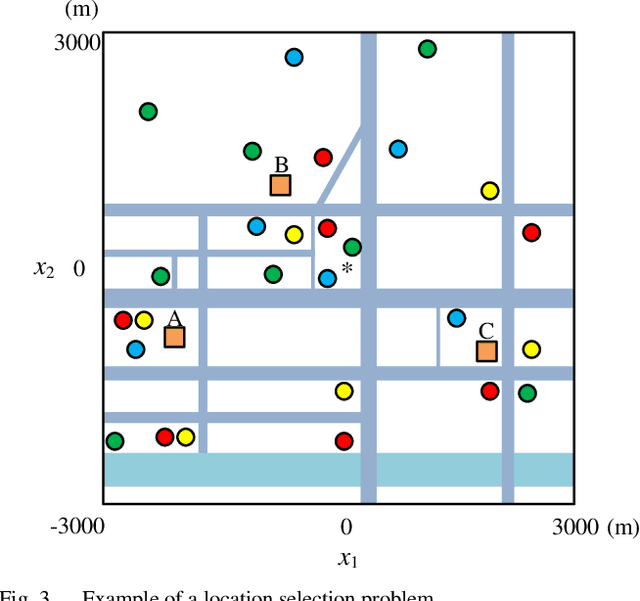

Abstract:In recent years, multimodal multiobjective optimization algorithms (MMOAs) based on evolutionary computation have been widely studied. However, existing MMOAs are mainly tested on benchmark function sets such as the 2019 IEEE Congress on Evolutionary Computation test suite (CEC 2019), and their performance on real-world problems is neglected. In this paper, two types of real-world multimodal multiobjective optimization problems in feature selection and location selection respectively are formulated. Moreover, four real-world datasets of Guangzhou, China are constructed for location selection. An investigation is conducted to evaluate the performance of seven existing MMOAs in solving these two types of real-world problems. An analysis of the experimental results explores the characteristics of the tested MMOAs, providing insights for selecting suitable MMOAs in real-world applications.
Privacy-Preserving Federated Foundation Model for Generalist Ultrasound Artificial Intelligence
Nov 25, 2024



Abstract:Ultrasound imaging is widely used in clinical diagnosis due to its non-invasive nature and real-time capabilities. However, conventional ultrasound diagnostics face several limitations, including high dependence on physician expertise and suboptimal image quality, which complicates interpretation and increases the likelihood of diagnostic errors. Artificial intelligence (AI) has emerged as a promising solution to enhance clinical diagnosis, particularly in detecting abnormalities across various biomedical imaging modalities. Nonetheless, current AI models for ultrasound imaging face critical challenges. First, these models often require large volumes of labeled medical data, raising concerns over patient privacy breaches. Second, most existing models are task-specific, which restricts their broader clinical utility. To overcome these challenges, we present UltraFedFM, an innovative privacy-preserving ultrasound foundation model. UltraFedFM is collaboratively pre-trained using federated learning across 16 distributed medical institutions in 9 countries, leveraging a dataset of over 1 million ultrasound images covering 19 organs and 10 ultrasound modalities. This extensive and diverse data, combined with a secure training framework, enables UltraFedFM to exhibit strong generalization and diagnostic capabilities. It achieves an average area under the receiver operating characteristic curve of 0.927 for disease diagnosis and a dice similarity coefficient of 0.878 for lesion segmentation. Notably, UltraFedFM surpasses the diagnostic accuracy of mid-level ultrasonographers and matches the performance of expert-level sonographers in the joint diagnosis of 8 common systemic diseases. These findings indicate that UltraFedFM can significantly enhance clinical diagnostics while safeguarding patient privacy, marking an advancement in AI-driven ultrasound imaging for future clinical applications.
A Simplifying and Learnable Graph Convolutional Attention Network for Unsupervised Knowledge Graphs Alignment
Oct 17, 2024



Abstract:The success of current Entity Alignment (EA) task depends largely on the supervision information provided by labeled data. Considering the cost of labeled data, most supervised methods are difficult to apply in practical scenarios. Therefore, more and more works based on contrastive learning, active learning or other deep learning techniques have been developed, to solve the performance bottleneck caused by the lack of labeled data. However, the existing unsupervised EA methods still have some limitations, either their modeling complexity is high or they cannot balance the effectiveness and practicality of alignment. To overcome these issues, we propose a Simplifying and Learnable graph convolutional attention network for Unsupervised Knowledge Graphs alignment method (SLU). Specifically, we first introduce LCAT, a new and simple framework as the backbone network to model the graph structure of two KGs. Then we design a reconstruction method of relation structure based on potential matching relations for efficiently filtering invalid neighborhood information of aligned entities, to improve the usability and scalability of SLU. Impressively, a similarity function based on consistency is proposed to better measure the similarity of candidate entity pairs. Finally, we conduct extensive experiments on three datasets of different sizes (15K and 100K) and different types (cross-lingual and monolingual) to verify the superiority of SLU. Experimental results show that SLU significantly improves alignment accuracy, outperforming 25 supervised or unsupervised methods, and improving 6.4% in Hits@1 over the best baseline in the best case.
MixPolyp: Integrating Mask, Box and Scribble Supervision for Enhanced Polyp Segmentation
Sep 25, 2024



Abstract:Limited by the expensive labeling, polyp segmentation models are plagued by data shortages. To tackle this, we propose the mixed supervised polyp segmentation paradigm (MixPolyp). Unlike traditional models relying on a single type of annotation, MixPolyp combines diverse annotation types (mask, box, and scribble) within a single model, thereby expanding the range of available data and reducing labeling costs. To achieve this, MixPolyp introduces three novel supervision losses to handle various annotations: Subspace Projection loss (L_SP), Binary Minimum Entropy loss (L_BME), and Linear Regularization loss (L_LR). For box annotations, L_SP eliminates shape inconsistencies between the prediction and the supervision. For scribble annotations, L_BME provides supervision for unlabeled pixels through minimum entropy constraint, thereby alleviating supervision sparsity. Furthermore, L_LR provides dense supervision by enforcing consistency among the predictions, thus reducing the non-uniqueness. These losses are independent of the model structure, making them generally applicable. They are used only during training, adding no computational cost during inference. Extensive experiments on five datasets demonstrate MixPolyp's effectiveness.
Let Video Teaches You More: Video-to-Image Knowledge Distillation using DEtection TRansformer for Medical Video Lesion Detection
Aug 26, 2024



Abstract:AI-assisted lesion detection models play a crucial role in the early screening of cancer. However, previous image-based models ignore the inter-frame contextual information present in videos. On the other hand, video-based models capture the inter-frame context but are computationally expensive. To mitigate this contradiction, we delve into Video-to-Image knowledge distillation leveraging DEtection TRansformer (V2I-DETR) for the task of medical video lesion detection. V2I-DETR adopts a teacher-student network paradigm. The teacher network aims at extracting temporal contexts from multiple frames and transferring them to the student network, and the student network is an image-based model dedicated to fast prediction in inference. By distilling multi-frame contexts into a single frame, the proposed V2I-DETR combines the advantages of utilizing temporal contexts from video-based models and the inference speed of image-based models. Through extensive experiments, V2I-DETR outperforms previous state-of-the-art methods by a large margin while achieving the real-time inference speed (30 FPS) as the image-based model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge