Yiwen Hu
SWE-World: Building Software Engineering Agents in Docker-Free Environments
Feb 03, 2026Abstract:Recent advances in large language models (LLMs) have enabled software engineering agents to tackle complex code modification tasks. Most existing approaches rely on execution feedback from containerized environments, which require dependency-complete setup and physical execution of programs and tests. While effective, this paradigm is resource-intensive and difficult to maintain, substantially complicating agent training and limiting scalability. We propose SWE-World, a Docker-free framework that replaces physical execution environments with a learned surrogate for training and evaluating software engineering agents. SWE-World leverages LLM-based models trained on real agent-environment interaction data to predict intermediate execution outcomes and final test feedback, enabling agents to learn without interacting with physical containerized environments. This design preserves the standard agent-environment interaction loop while eliminating the need for costly environment construction and maintenance during agent optimization and evaluation. Furthermore, because SWE-World can simulate the final evaluation outcomes of candidate trajectories without real submission, it enables selecting the best solution among multiple test-time attempts, thereby facilitating effective test-time scaling (TTS) in software engineering tasks. Experiments on SWE-bench Verified demonstrate that SWE-World raises Qwen2.5-Coder-32B from 6.2\% to 52.0\% via Docker-free SFT, 55.0\% with Docker-free RL, and 68.2\% with further TTS. The code is available at https://github.com/RUCAIBox/SWE-World
SWE-Master: Unleashing the Potential of Software Engineering Agents via Post-Training
Feb 03, 2026Abstract:In this technical report, we present SWE-Master, an open-source and fully reproducible post-training framework for building effective software engineering agents. SWE-Master systematically explores the complete agent development pipeline, including teacher-trajectory synthesis and data curation, long-horizon SFT, RL with real execution feedback, and inference framework design. Starting from an open-source base model with limited initial SWE capability, SWE-Master demonstrates how systematical optimization method can elicit strong long-horizon SWE task solving abilities. We evaluate SWE-Master on SWE-bench Verified, a standard benchmark for realistic software engineering tasks. Under identical experimental settings, our approach achieves a resolve rate of 61.4\% with Qwen2.5-Coder-32B, substantially outperforming existing open-source baselines. By further incorporating test-time scaling~(TTS) with LLM-based environment feedback, SWE-Master reaches 70.8\% at TTS@8, demonstrating a strong performance potential. SWE-Master provides a practical and transparent foundation for advancing reproducible research on software engineering agents. The code is available at https://github.com/RUCAIBox/SWE-Master.
Controlled LLM Training on Spectral Sphere
Jan 13, 2026Abstract:Scaling large models requires optimization strategies that ensure rapid convergence grounded in stability. Maximal Update Parametrization ($\boldsymbolμ$P) provides a theoretical safeguard for width-invariant $Θ(1)$ activation control, whereas emerging optimizers like Muon are only ``half-aligned'' with these constraints: they control updates but allow weights to drift. To address this limitation, we introduce the \textbf{Spectral Sphere Optimizer (SSO)}, which enforces strict module-wise spectral constraints on both weights and their updates. By deriving the steepest descent direction on the spectral sphere, SSO realizes a fully $\boldsymbolμ$P-aligned optimization process. To enable large-scale training, we implement SSO as an efficient parallel algorithm within Megatron. Through extensive pretraining on diverse architectures, including Dense 1.7B, MoE 8B-A1B, and 200-layer DeepNet models, SSO consistently outperforms AdamW and Muon. Furthermore, we observe significant practical stability benefits, including improved MoE router load balancing, suppressed outliers, and strictly bounded activations.
Entropy-Guided Token Dropout: Training Autoregressive Language Models with Limited Domain Data
Dec 29, 2025Abstract:As access to high-quality, domain-specific data grows increasingly scarce, multi-epoch training has become a practical strategy for adapting large language models (LLMs). However, autoregressive models often suffer from performance degradation under repeated data exposure, where overfitting leads to a marked decline in model capability. Through empirical analysis, we trace this degradation to an imbalance in learning dynamics: predictable, low-entropy tokens are learned quickly and come to dominate optimization, while the model's ability to generalize on high-entropy tokens deteriorates with continued training. To address this, we introduce EntroDrop, an entropy-guided token dropout method that functions as structured data regularization. EntroDrop selectively masks low-entropy tokens during training and employs a curriculum schedule to adjust regularization strength in alignment with training progress. Experiments across model scales from 0.6B to 8B parameters show that EntroDrop consistently outperforms standard regularization baselines and maintains robust performance throughout extended multi-epoch training. These findings underscore the importance of aligning regularization with token-level learning dynamics when training on limited data. Our approach offers a promising pathway toward more effective adaptation of LLMs in data-constrained domains.
ReaSeq: Unleashing World Knowledge via Reasoning for Sequential Modeling
Dec 24, 2025Abstract:Industrial recommender systems face two fundamental limitations under the log-driven paradigm: (1) knowledge poverty in ID-based item representations that causes brittle interest modeling under data sparsity, and (2) systemic blindness to beyond-log user interests that constrains model performance within platform boundaries. These limitations stem from an over-reliance on shallow interaction statistics and close-looped feedback while neglecting the rich world knowledge about product semantics and cross-domain behavioral patterns that Large Language Models have learned from vast corpora. To address these challenges, we introduce ReaSeq, a reasoning-enhanced framework that leverages world knowledge in Large Language Models to address both limitations through explicit and implicit reasoning. Specifically, ReaSeq employs explicit Chain-of-Thought reasoning via multi-agent collaboration to distill structured product knowledge into semantically enriched item representations, and latent reasoning via Diffusion Large Language Models to infer plausible beyond-log behaviors. Deployed on Taobao's ranking system serving hundreds of millions of users, ReaSeq achieves substantial gains: >6.0% in IPV and CTR, >2.9% in Orders, and >2.5% in GMV, validating the effectiveness of world-knowledge-enhanced reasoning over purely log-driven approaches.
A FEDformer-Based Hybrid Framework for Anomaly Detection and Risk Forecasting in Financial Time Series
Nov 17, 2025Abstract:Financial markets are inherently volatile and prone to sudden disruptions such as market crashes, flash collapses, and liquidity crises. Accurate anomaly detection and early risk forecasting in financial time series are therefore crucial for preventing systemic instability and supporting informed investment decisions. Traditional deep learning models, such as LSTM and GRU, often fail to capture long-term dependencies and complex periodic patterns in highly nonstationary financial data. To address this limitation, this study proposes a FEDformer-Based Hybrid Framework for Anomaly Detection and Risk Forecasting in Financial Time Series, which integrates the Frequency Enhanced Decomposed Transformer (FEDformer) with a residual-based anomaly detector and a risk forecasting head. The FEDformer module models temporal dynamics in both time and frequency domains, decomposing signals into trend and seasonal components for improved interpretability. The residual-based detector identifies abnormal fluctuations by analyzing prediction errors, while the risk head predicts potential financial distress using learned latent embeddings. Experiments conducted on the S&P 500, NASDAQ Composite, and Brent Crude Oil datasets (2000-2024) demonstrate the superiority of the proposed model over benchmark methods, achieving a 15.7 percent reduction in RMSE and an 11.5 percent improvement in F1-score for anomaly detection. These results confirm the effectiveness of the model in capturing financial volatility, enabling reliable early-warning systems for market crash prediction and risk management.
ICPC-Eval: Probing the Frontiers of LLM Reasoning with Competitive Programming Contests
Jun 05, 2025Abstract:With the significant progress of large reasoning models in complex coding and reasoning tasks, existing benchmarks, like LiveCodeBench and CodeElo, are insufficient to evaluate the coding capabilities of large language models (LLMs) in real competition environments. Moreover, current evaluation metrics such as Pass@K fail to capture the reflective abilities of reasoning models. To address these challenges, we propose \textbf{ICPC-Eval}, a top-level competitive coding benchmark designed to probing the frontiers of LLM reasoning. ICPC-Eval includes 118 carefully curated problems from 11 recent ICPC contests held in various regions of the world, offering three key contributions: 1) A challenging realistic ICPC competition scenario, featuring a problem type and difficulty distribution consistent with actual contests. 2) A robust test case generation method and a corresponding local evaluation toolkit, enabling efficient and accurate local evaluation. 3) An effective test-time scaling evaluation metric, Refine@K, which allows iterative repair of solutions based on execution feedback. The results underscore the significant challenge in evaluating complex reasoning abilities: top-tier reasoning models like DeepSeek-R1 often rely on multi-turn code feedback to fully unlock their in-context reasoning potential when compared to non-reasoning counterparts. Furthermore, despite recent advancements in code generation, these models still lag behind top-performing human teams. We release the benchmark at: https://github.com/RUCAIBox/Slow_Thinking_with_LLMs
MARIO: A Mixed Annotation Framework For Polyp Segmentation
Jan 19, 2025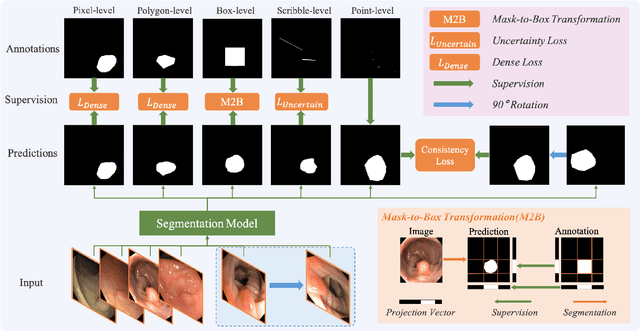
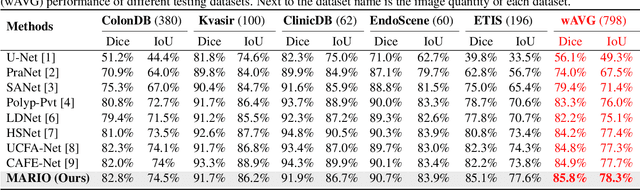
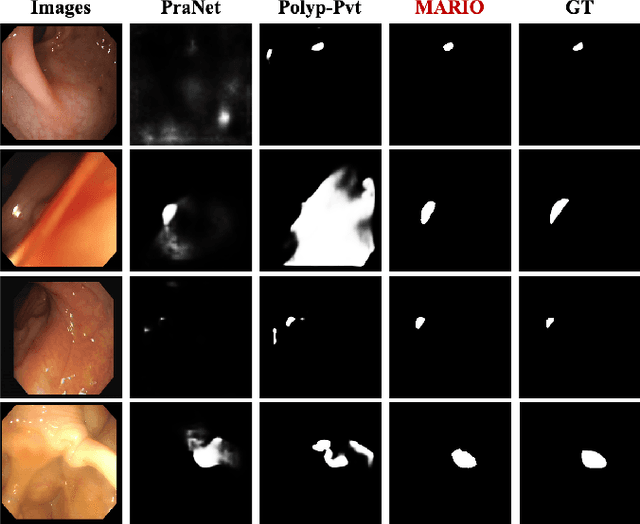

Abstract:Existing polyp segmentation models are limited by high labeling costs and the small size of datasets. Additionally, vast polyp datasets remain underutilized because these models typically rely on a single type of annotation. To address this dilemma, we introduce MARIO, a mixed supervision model designed to accommodate various annotation types, significantly expanding the range of usable data. MARIO learns from underutilized datasets by incorporating five forms of supervision: pixel-level, box-level, polygon-level, scribblelevel, and point-level. Each form of supervision is associated with a tailored loss that effectively leverages the supervision labels while minimizing the noise. This allows MARIO to move beyond the constraints of relying on a single annotation type. Furthermore, MARIO primarily utilizes dataset with weak and cheap annotations, reducing the dependence on large-scale, fully annotated ones. Experimental results across five benchmark datasets demonstrate that MARIO consistently outperforms existing methods, highlighting its efficacy in balancing trade-offs between different forms of supervision and maximizing polyp segmentation performance
YuLan-Mini: An Open Data-efficient Language Model
Dec 24, 2024



Abstract:Effective pre-training of large language models (LLMs) has been challenging due to the immense resource demands and the complexity of the technical processes involved. This paper presents a detailed technical report on YuLan-Mini, a highly capable base model with 2.42B parameters that achieves top-tier performance among models of similar parameter scale. Our pre-training approach focuses on enhancing training efficacy through three key technical contributions: an elaborate data pipeline combines data cleaning with data schedule strategies, a robust optimization method to mitigate training instability, and an effective annealing approach that incorporates targeted data selection and long context training. Remarkably, YuLan-Mini, trained on 1.08T tokens, achieves performance comparable to industry-leading models that require significantly more data. To facilitate reproduction, we release the full details of the data composition for each training phase. Project details can be accessed at the following link: https://github.com/RUC-GSAI/YuLan-Mini.
Imitate, Explore, and Self-Improve: A Reproduction Report on Slow-thinking Reasoning Systems
Dec 12, 2024


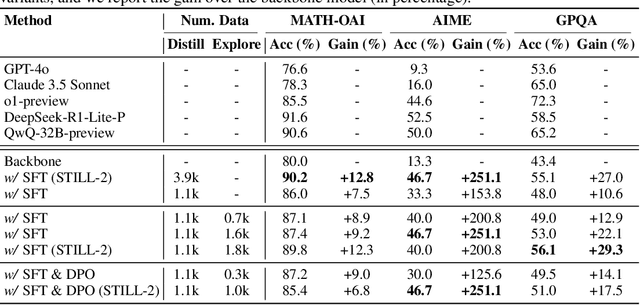
Abstract:Recently, slow-thinking reasoning systems, such as o1, have demonstrated remarkable capabilities in solving complex reasoning tasks. These systems typically engage in an extended thinking process before responding to a query, allowing them to generate more thorough, accurate, and well-reasoned solutions. These systems are primarily developed and maintained by industry, with their core techniques not publicly disclosed. In response, an increasing number of studies from the research community aim to explore the technical foundations underlying these powerful reasoning systems. Building on these prior efforts, this paper presents a reproduction report on implementing o1-like reasoning systems. We introduce an "imitate, explore, and self-improve" framework as our primary technical approach to train the reasoning model. In the initial phase, we use distilled long-form thought data to fine-tune the reasoning model, enabling it to invoke a slow-thinking mode. The model is then encouraged to explore challenging problems by generating multiple rollouts, which can result in increasingly more high-quality trajectories that lead to correct answers. Furthermore, the model undergoes self-improvement by iteratively refining its training dataset. To verify the effectiveness of this approach, we conduct extensive experiments on three challenging benchmarks. The experimental results demonstrate that our approach achieves competitive performance compared to industry-level reasoning systems on these benchmarks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge