Sumei Fan
ECGomics: An Open Platform for AI-ECG Digital Biomarker Discovery
Jan 19, 2026Abstract:Background: Conventional electrocardiogram (ECG) analysis faces a persistent dichotomy: expert-driven features ensure interpretability but lack sensitivity to latent patterns, while deep learning offers high accuracy but functions as a black box with high data dependency. We introduce ECGomics, a systematic paradigm and open-source platform for the multidimensional deconstruction of cardiac signals into digital biomarker. Methods: Inspired by the taxonomic rigor of genomics, ECGomics deconstructs cardiac activity across four dimensions: Structural, Intensity, Functional, and Comparative. This taxonomy synergizes expert-defined morphological rules with data-driven latent representations, effectively bridging the gap between handcrafted features and deep learning embeddings. Results: We operationalized this framework into a scalable ecosystem consisting of a web-based research platform and a mobile-integrated solution (https://github.com/PKUDigitalHealth/ECGomics). The web platform facilitates high-throughput analysis via precision parameter configuration, high-fidelity data ingestion, and 12-lead visualization, allowing for the systematic extraction of biomarkers across the four ECGomics dimensions. Complementarily, the mobile interface, integrated with portable sensors and a cloud-based engine, enables real-time signal acquisition and near-instantaneous delivery of structured diagnostic reports. This dual-interface architecture successfully transitions ECGomics from theoretical discovery to decentralized, real-world health management, ensuring professional-grade monitoring in diverse clinical and home-based settings. Conclusion: ECGomics harmonizes diagnostic precision, interpretability, and data efficiency. By providing a deployable software ecosystem, this paradigm establishes a robust foundation for digital biomarker discovery and personalized cardiovascular medicine.
Accuracy of Wearable ECG Parameter Calculation Method for Long QT and First-Degree A-V Block Detection: A Multi-Center Real-World Study with External Validations Compared to Standard ECG Machines and Cardiologist Assessments
Feb 21, 2025Abstract:In recent years, wearable devices have revolutionized cardiac monitoring by enabling continuous, non-invasive ECG recording in real-world settings. Despite these advances, the accuracy of ECG parameter calculations (PR interval, QRS interval, QT interval, etc.) from wearables remains to be rigorously validated against conventional ECG machines and expert clinician assessments. In this large-scale, multicenter study, we evaluated FeatureDB, a novel algorithm for automated computation of ECG parameters from wearable single-lead signals Three diverse datasets were employed: the AHMU-FH dataset (n=88,874), the CSE dataset (n=106), and the HeartVoice-ECG-lite dataset (n=369) with annotations provided by two experienced cardiologists. FeatureDB demonstrates a statistically significant correlation with key parameters (PR interval, QRS duration, QT interval, and QTc) calculated by standard ECG machines and annotated by clinical doctors. Bland-Altman analysis confirms a high level of agreement.Moreover,FeatureDB exhibited robust diagnostic performance in detecting Long QT syndrome (LQT) and atrioventricular block interval abnormalities (AVBI),with excellent area under the ROC curve (LQT: 0.836, AVBI: 0.861),accuracy (LQT: 0.856, AVBI: 0.845),sensitivity (LQT: 0.815, AVBI: 0.877),and specificity (LQT: 0.856, AVBI: 0.845).This further validates its clinical reliability. These results validate the clinical applicability of FeatureDB for wearable ECG analysis and highlight its potential to bridge the gap between traditional diagnostic methods and emerging wearable technologies.Ultimately,this study supports integrating wearable ECG devices into large-scale cardiovascular disease management and early intervention strategies,and it highlights the potential of wearable ECG technologies to deliver accurate,clinically relevant cardiac monitoring while advancing broader applications in cardiovascular care.
Artificial Intelligence System for Detection and Screening of Cardiac Abnormalities using Electrocardiogram Images
Feb 10, 2023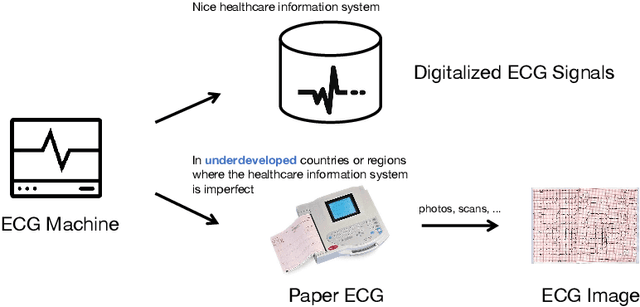



Abstract:The artificial intelligence (AI) system has achieved expert-level performance in electrocardiogram (ECG) signal analysis. However, in underdeveloped countries or regions where the healthcare information system is imperfect, only paper ECGs can be provided. Analysis of real-world ECG images (photos or scans of paper ECGs) remains challenging due to complex environments or interference. In this study, we present an AI system developed to detect and screen cardiac abnormalities (CAs) from real-world ECG images. The system was evaluated on a large dataset of 52,357 patients from multiple regions and populations across the world. On the detection task, the AI system obtained area under the receiver operating curve (AUC) of 0.996 (hold-out test), 0.994 (external test 1), 0.984 (external test 2), and 0.979 (external test 3), respectively. Meanwhile, the detection results of AI system showed a strong correlation with the diagnosis of cardiologists (cardiologist 1 (R=0.794, p<1e-3), cardiologist 2 (R=0.812, p<1e-3)). On the screening task, the AI system achieved AUCs of 0.894 (hold-out test) and 0.850 (external test). The screening performance of the AI system was better than that of the cardiologists (AI system (0.846) vs. cardiologist 1 (0.520) vs. cardiologist 2 (0.480)). Our study demonstrates the feasibility of an accurate, objective, easy-to-use, fast, and low-cost AI system for CA detection and screening. The system has the potential to be used by healthcare professionals, caregivers, and general users to assess CAs based on real-world ECG images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge