Guodong Wei
Artificial Intelligence System for Detection and Screening of Cardiac Abnormalities using Electrocardiogram Images
Feb 10, 2023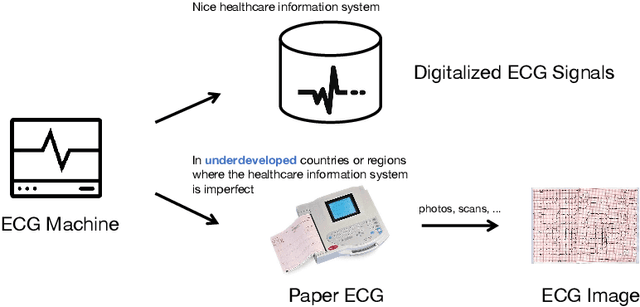



Abstract:The artificial intelligence (AI) system has achieved expert-level performance in electrocardiogram (ECG) signal analysis. However, in underdeveloped countries or regions where the healthcare information system is imperfect, only paper ECGs can be provided. Analysis of real-world ECG images (photos or scans of paper ECGs) remains challenging due to complex environments or interference. In this study, we present an AI system developed to detect and screen cardiac abnormalities (CAs) from real-world ECG images. The system was evaluated on a large dataset of 52,357 patients from multiple regions and populations across the world. On the detection task, the AI system obtained area under the receiver operating curve (AUC) of 0.996 (hold-out test), 0.994 (external test 1), 0.984 (external test 2), and 0.979 (external test 3), respectively. Meanwhile, the detection results of AI system showed a strong correlation with the diagnosis of cardiologists (cardiologist 1 (R=0.794, p<1e-3), cardiologist 2 (R=0.812, p<1e-3)). On the screening task, the AI system achieved AUCs of 0.894 (hold-out test) and 0.850 (external test). The screening performance of the AI system was better than that of the cardiologists (AI system (0.846) vs. cardiologist 1 (0.520) vs. cardiologist 2 (0.480)). Our study demonstrates the feasibility of an accurate, objective, easy-to-use, fast, and low-cost AI system for CA detection and screening. The system has the potential to be used by healthcare professionals, caregivers, and general users to assess CAs based on real-world ECG images.
A Deep Bayesian Neural Network for Cardiac Arrhythmia Classification with Rejection from ECG Recordings
Feb 26, 2022

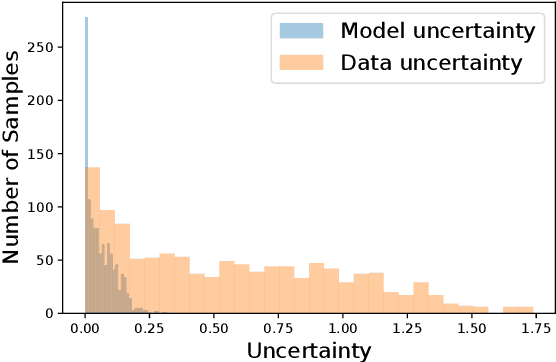
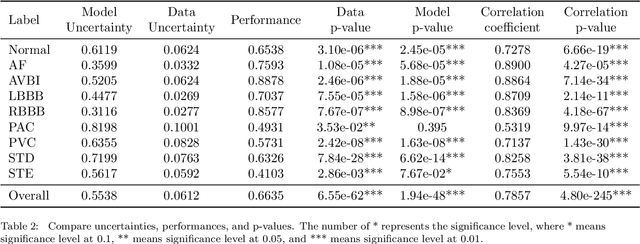
Abstract:With the development of deep learning-based methods, automated classification of electrocardiograms (ECGs) has recently gained much attention. Although the effectiveness of deep neural networks has been encouraging, the lack of information given by the outputs restricts clinicians' reexamination. If the uncertainty estimation comes along with the classification results, cardiologists can pay more attention to "uncertain" cases. Our study aims to classify ECGs with rejection based on data uncertainty and model uncertainty. We perform experiments on a real-world 12-lead ECG dataset. First, we estimate uncertainties using the Monte Carlo dropout for each classification prediction, based on our Bayesian neural network. Then, we accept predictions with uncertainty under a given threshold and provide "uncertain" cases for clinicians. Furthermore, we perform a simulation experiment using varying thresholds. Finally, with the help of a clinician, we conduct case studies to explain the results of large uncertainties and incorrect predictions with small uncertainties. The results show that correct predictions are more likely to have smaller uncertainties, and the performance on accepted predictions improves as the accepting ratio decreases (i.e. more rejections). Case studies also help explain why rejection can improve the performance. Our study helps neural networks produce more accurate results and provide information on uncertainties to better assist clinicians in the diagnosis process. It can also enable deep-learning-based ECG interpretation in clinical implementation.
Semi-supervised Anatomical Landmark Detection via Shape-regulated Self-training
May 28, 2021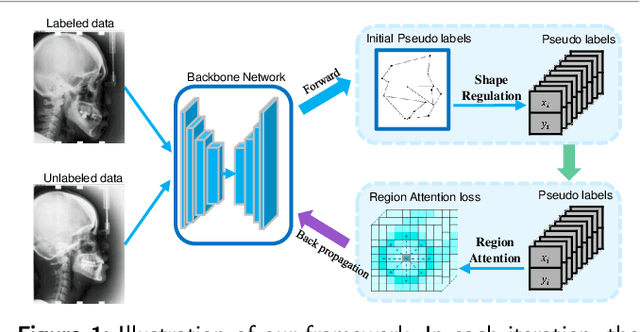
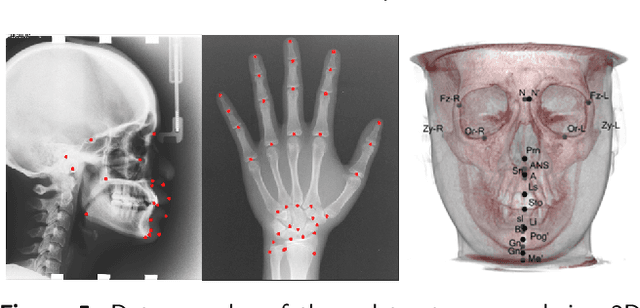

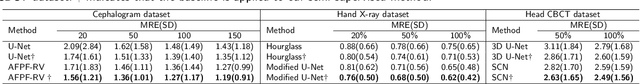
Abstract:Well-annotated medical images are costly and sometimes even impossible to acquire, hindering landmark detection accuracy to some extent. Semi-supervised learning alleviates the reliance on large-scale annotated data by exploiting the unlabeled data to understand the population structure of anatomical landmarks. The global shape constraint is the inherent property of anatomical landmarks that provides valuable guidance for more consistent pseudo labelling of the unlabeled data, which is ignored in the previously semi-supervised methods. In this paper, we propose a model-agnostic shape-regulated self-training framework for semi-supervised landmark detection by fully considering the global shape constraint. Specifically, to ensure pseudo labels are reliable and consistent, a PCA-based shape model adjusts pseudo labels and eliminate abnormal ones. A novel Region Attention loss to make the network automatically focus on the structure consistent regions around pseudo labels. Extensive experiments show that our approach outperforms other semi-supervised methods and achieves notable improvement on three medical image datasets. Moreover, our framework is flexible and can be used as a plug-and-play module integrated into most supervised methods to improve performance further.
Attending Category Disentangled Global Context for Image Classification
Dec 17, 2018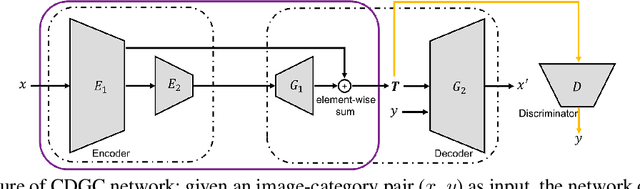
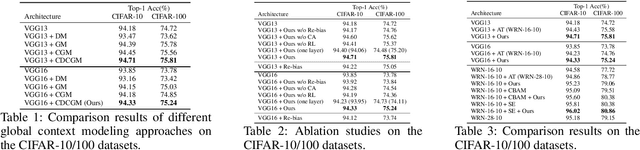
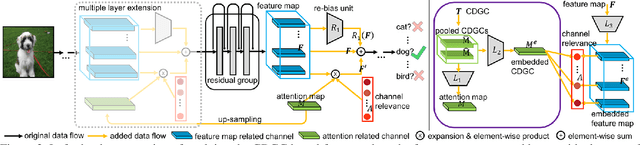
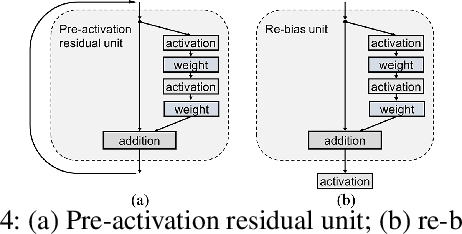
Abstract:In this paper, we propose a general framework for image classification using the attention mechanism and global context, which could incorporate with various network architectures to improve their performance. To investigate the capability of the global context, we compare four mathematical models and observe the global context encoded in the category disentangled conditional generative model retains the richest complementary information to that in the baseline classification networks. Based on this observation, we define a novel Category Disentangled Global Context (CDGC) and devise a deep network to obtain it. By attending CDGC, the baseline networks could identify the objects of interest more accurately, thus improving the performance. We apply the framework to many different network architectures to demonstrate its effectiveness and versatility. Extensive results on four publicly available datasets validate our approach could generalize well and is superior to the state-of-the-art. In addition, the framework could be combined with various self-attention based methods to further promote the performance. Code and pretrained models will be made public upon paper acceptance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge