Siddharth Biswal
Automated Respiratory Event Detection Using Deep Neural Networks
Jan 12, 2021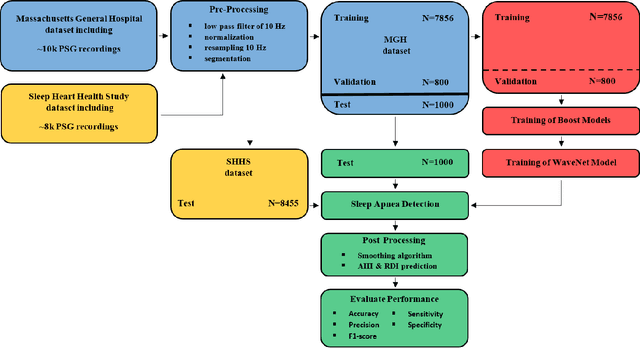
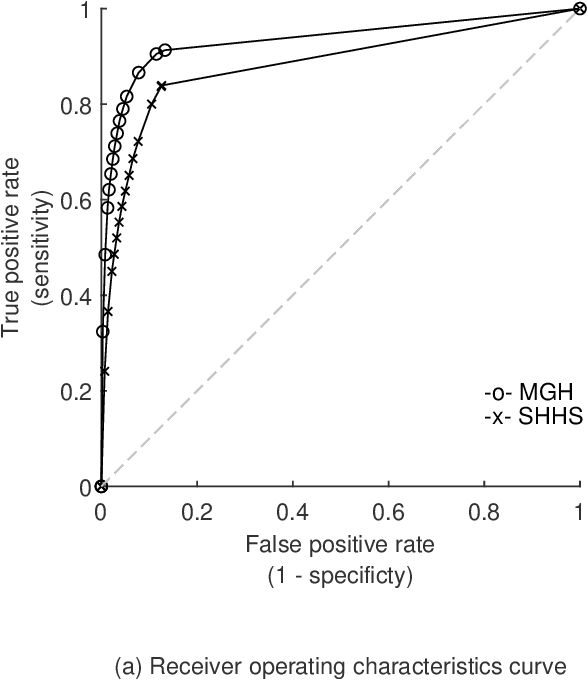
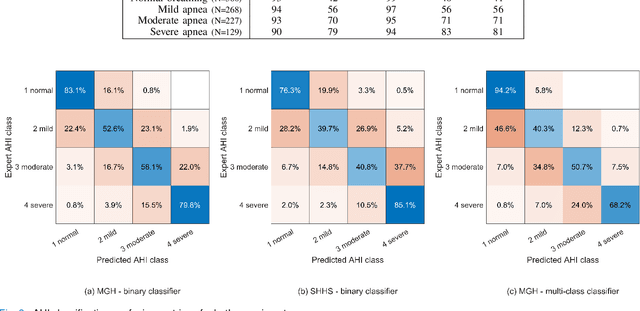
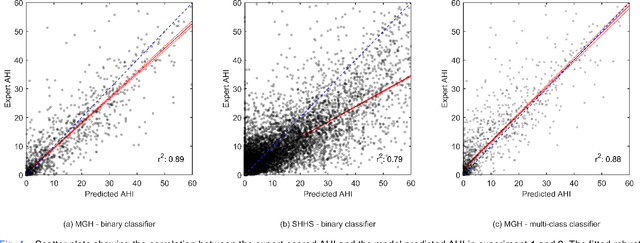
Abstract:The gold standard to assess respiration during sleep is polysomnography; a technique that is burdensome, expensive (both in analysis time and measurement costs), and difficult to repeat. Automation of respiratory analysis can improve test efficiency and enable accessible implementation opportunities worldwide. Using 9,656 polysomnography recordings from the Massachusetts General Hospital (MGH), we trained a neural network (WaveNet) based on a single respiratory effort belt to detect obstructive apnea, central apnea, hypopnea and respiratory-effort related arousals. Performance evaluation included event-based and recording-based metrics - using an apnea-hypopnea index analysis. The model was further evaluated on a public dataset, the Sleep-Heart-Health-Study-1, containing 8,455 polysomnographic recordings. For binary apnea event detection in the MGH dataset, the neural network obtained an accuracy of 95%, an apnea-hypopnea index $r^2$ of 0.89 and area under the curve for the receiver operating characteristics curve and precision-recall curve of 0.93 and 0.74, respectively. For the multiclass task, we obtained varying performances: 81% of all labeled central apneas were correctly classified, whereas this metric was 46% for obstructive apneas, 29% for respiratory effort related arousals and 16% for hypopneas. The majority of false predictions were misclassifications as another type of respiratory event. Our fully automated method can detect respiratory events and assess the apnea-hypopnea index with sufficient accuracy for clinical utilization. Differentiation of event types is more difficult and may reflect in part the complexity of human respiratory output and some degree of arbitrariness in the clinical thresholds and criteria used during manual annotation.
EVA: Generating Longitudinal Electronic Health Records Using Conditional Variational Autoencoders
Dec 18, 2020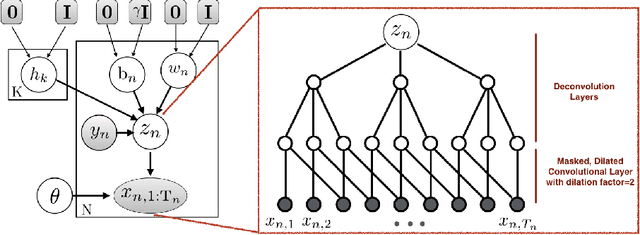
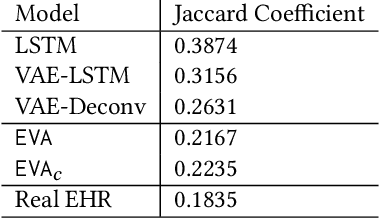
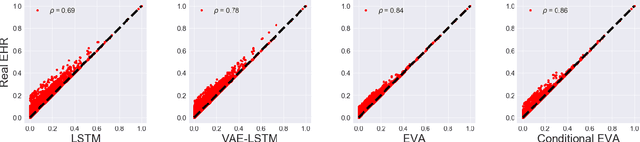
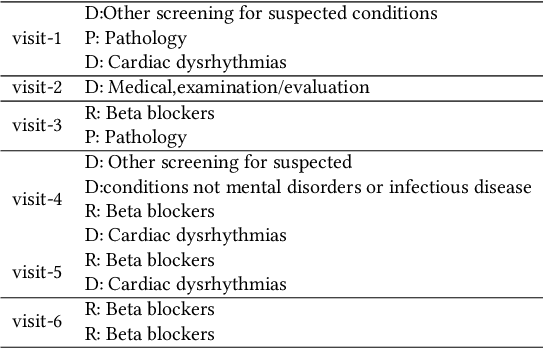
Abstract:Researchers require timely access to real-world longitudinal electronic health records (EHR) to develop, test, validate, and implement machine learning solutions that improve the quality and efficiency of healthcare. In contrast, health systems value deeply patient privacy and data security. De-identified EHRs do not adequately address the needs of health systems, as de-identified data are susceptible to re-identification and its volume is also limited. Synthetic EHRs offer a potential solution. In this paper, we propose EHR Variational Autoencoder (EVA) for synthesizing sequences of discrete EHR encounters (e.g., clinical visits) and encounter features (e.g., diagnoses, medications, procedures). We illustrate that EVA can produce realistic EHR sequences, account for individual differences among patients, and can be conditioned on specific disease conditions, thus enabling disease-specific studies. We design efficient, accurate inference algorithms by combining stochastic gradient Markov Chain Monte Carlo with amortized variational inference. We assess the utility of the methods on large real-world EHR repositories containing over 250, 000 patients. Our experiments, which include user studies with knowledgeable clinicians, indicate the generated EHR sequences are realistic. We confirmed the performance of predictive models trained on the synthetic data are similar with those trained on real EHRs. Additionally, our findings indicate that augmenting real data with synthetic EHRs results in the best predictive performance - improving the best baseline by as much as 8% in top-20 recall.
EMIXER: End-to-end Multimodal X-ray Generation via Self-supervision
Jul 10, 2020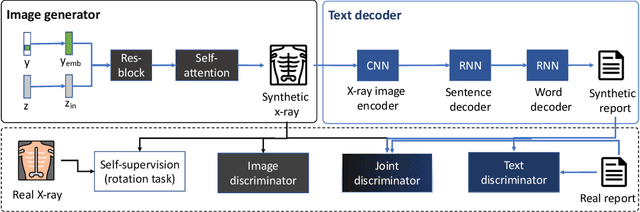

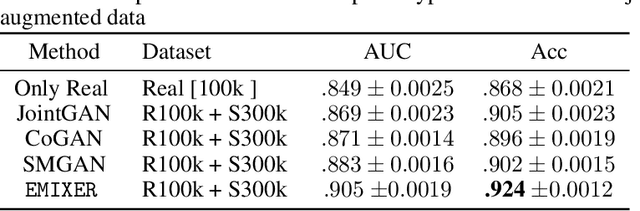

Abstract:Deep generative models have enabled the automated synthesis of high-quality data for diverse applications. However, the most effective generative models are specialized to data from a single domain (e.g., images or text). Real-world applications such as healthcare require multi-modal data from multiple domains (e.g., both images and corresponding text), which are difficult to acquire due to limited availability and privacy concerns and are much harder to synthesize. To tackle this joint synthesis challenge, we propose an End-to-end MultImodal X-ray genERative model (EMIXER) for jointly synthesizing x-ray images and corresponding free-text reports, all conditional on diagnosis labels. EMIXER is an conditional generative adversarial model by 1) generating an image based on a label, 2) encoding the image to a hidden embedding, 3) producing the corresponding text via a hierarchical decoder from the image embedding, and 4) a joint discriminator for assessing both the image and the corresponding text. EMIXER also enables self-supervision to leverage vast amount of unlabeled data. Extensive experiments with real X-ray reports data illustrate how data augmentation using synthesized multimodal samples can improve the performance of a variety of supervised tasks including COVID-19 X-ray classification with very limited samples. The quality of generated images and reports are also confirmed by radiologists. We quantitatively show that EMIXER generated synthetic datasets can augment X-ray image classification, report generation models to achieve 5.94% and 6.9% improvement on models trained only on real data samples. Taken together, our results highlight the promise of state of generative models to advance clinical machine learning.
CLARA: Clinical Report Auto-completion
Mar 04, 2020



Abstract:Generating clinical reports from raw recordings such as X-rays and electroencephalogram (EEG) is an essential and routine task for doctors. However, it is often time-consuming to write accurate and detailed reports. Most existing methods try to generate the whole reports from the raw input with limited success because 1) generated reports often contain errors that need manual review and correction, 2) it does not save time when doctors want to write additional information into the report, and 3) the generated reports are not customized based on individual doctors' preference. We propose {\it CL}inic{\it A}l {\it R}eport {\it A}uto-completion (CLARA), an interactive method that generates reports in a sentence by sentence fashion based on doctors' anchor words and partially completed sentences. CLARA searches for most relevant sentences from existing reports as the template for the current report. The retrieved sentences are sequentially modified by combining with the input feature representations to create the final report. In our experimental evaluation, CLARA achieved 0.393 CIDEr and 0.248 BLEU-4 on X-ray reports and 0.482 CIDEr and 0.491 BLEU-4 for EEG reports for sentence-level generation, which is up to 35% improvement over the best baseline. Also via our qualitative evaluation, CLARA is shown to produce reports which have a significantly higher level of approval by doctors in a user study (3.74 out of 5 for CLARA vs 2.52 out of 5 for the baseline).
CONAN: Complementary Pattern Augmentation for Rare Disease Detection
Nov 26, 2019



Abstract:Rare diseases affect hundreds of millions of people worldwide but are hard to detect since they have extremely low prevalence rates (varying from 1/1,000 to 1/200,000 patients) and are massively underdiagnosed. How do we reliably detect rare diseases with such low prevalence rates? How to further leverage patients with possibly uncertain diagnosis to improve detection? In this paper, we propose a Complementary pattern Augmentation (CONAN) framework for rare disease detection. CONAN combines ideas from both adversarial training and max-margin classification. It first learns self-attentive and hierarchical embedding for patient pattern characterization. Then, we develop a complementary generative adversarial networks (GAN) model to generate candidate positive and negative samples from the uncertain patients by encouraging a max-margin between classes. In addition, CONAN has a disease detector that serves as the discriminator during the adversarial training for identifying rare diseases. We evaluated CONAN on two disease detection tasks. For low prevalence inflammatory bowel disease (IBD) detection, CONAN achieved .96 precision recall area under the curve (PR-AUC) and 50.1% relative improvement over best baseline. For rare disease idiopathic pulmonary fibrosis (IPF) detection, CONAN achieves .22 PR-AUC with 41.3% relative improvement over the best baseline.
Doctor2Vec: Dynamic Doctor Representation Learning for Clinical Trial Recruitment
Nov 23, 2019



Abstract:Massive electronic health records (EHRs) enable the success of learning accurate patient representations to support various predictive health applications. In contrast, doctor representation was not well studied despite that doctors play pivotal roles in healthcare. How to construct the right doctor representations? How to use doctor representation to solve important health analytic problems? In this work, we study the problem on {\it clinical trial recruitment}, which is about identifying the right doctors to help conduct the trials based on the trial description and patient EHR data of those doctors. We propose doctor2vec which simultaneously learns 1) doctor representations from EHR data and 2) trial representations from the description and categorical information about the trials. In particular, doctor2vec utilizes a dynamic memory network where the doctor's experience with patients are stored in the memory bank and the network will dynamically assign weights based on the trial representation via an attention mechanism. Validated on large real-world trials and EHR data including 2,609 trials, 25K doctors and 430K patients, doctor2vec demonstrated improved performance over the best baseline by up to $8.7\%$ in PR-AUC. We also demonstrated that the doctor2vec embedding can be transferred to benefit data insufficiency settings including trial recruitment in less populated/newly explored country with $13.7\%$ improvement or for rare diseases with $8.1\%$ improvement in PR-AUC.
HAMLET: Interpretable Human And Machine co-LEarning Technique
Aug 21, 2018



Abstract:Efficient label acquisition processes are key to obtaining robust classifiers. However, data labeling is often challenging and subject to high levels of label noise. This can arise even when classification targets are well defined, if instances to be labeled are more difficult than the prototypes used to define the class, leading to disagreements among the expert community. Here, we enable efficient training of deep neural networks. From low-confidence labels, we iteratively improve their quality by simultaneous learning of machines and experts. We call it Human And Machine co-LEarning Technique (HAMLET). Throughout the process, experts become more consistent, while the algorithm provides them with explainable feedback for confirmation. HAMLET uses a neural embedding function and a memory module filled with diverse reference embeddings from different classes. Its output includes classification labels and highly relevant reference embeddings as explanation. We took the study of brain monitoring at intensive care unit (ICU) as an application of HAMLET on continuous electroencephalography (cEEG) data. Although cEEG monitoring yields large volumes of data, labeling costs and difficulty make it hard to build a classifier. Additionally, while experts agree on the labels of clear-cut examples of cEEG patterns, labeling many real-world cEEG data can be extremely challenging. Thus, a large minority of sequences might be mislabeled. HAMLET has shown significant performance gain against deep learning and other baselines, increasing accuracy from 7.03% to 68.75% on challenging inputs. Besides improved performance, clinical experts confirmed the interpretability of those reference embeddings in helping explaining the classification results by HAMLET.
RAIM: Recurrent Attentive and Intensive Model of Multimodal Patient Monitoring Data
Jul 23, 2018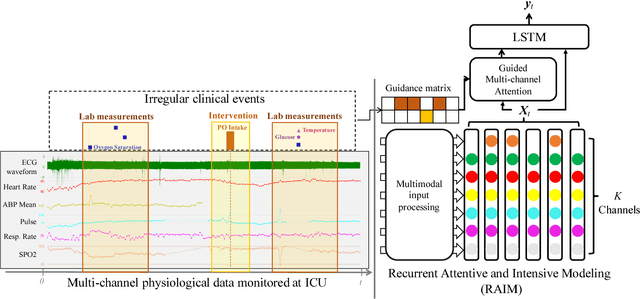

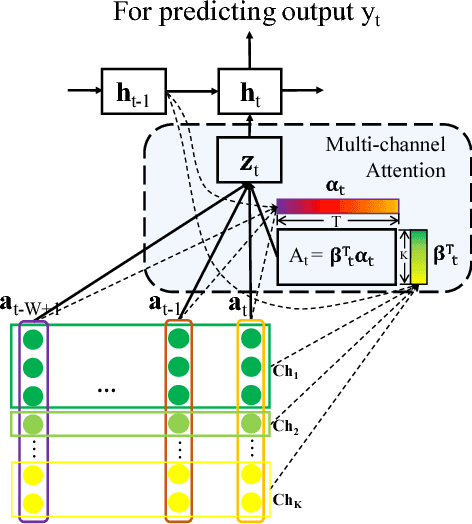

Abstract:With the improvement of medical data capturing, vast amount of continuous patient monitoring data, e.g., electrocardiogram (ECG), real-time vital signs and medications, become available for clinical decision support at intensive care units (ICUs). However, it becomes increasingly challenging to model such data, due to high density of the monitoring data, heterogeneous data types and the requirement for interpretable models. Integration of these high-density monitoring data with the discrete clinical events (including diagnosis, medications, labs) is challenging but potentially rewarding since richness and granularity in such multimodal data increase the possibilities for accurate detection of complex problems and predicting outcomes (e.g., length of stay and mortality). We propose Recurrent Attentive and Intensive Model (RAIM) for jointly analyzing continuous monitoring data and discrete clinical events. RAIM introduces an efficient attention mechanism for continuous monitoring data (e.g., ECG), which is guided by discrete clinical events (e.g, medication usage). We apply RAIM in predicting physiological decompensation and length of stay in those critically ill patients at ICU. With evaluations on MIMIC- III Waveform Database Matched Subset, we obtain an AUC-ROC score of 90.18% for predicting decompensation and an accuracy of 86.82% for forecasting length of stay with our final model, which outperforms our six baseline models.
Generating Multi-label Discrete Patient Records using Generative Adversarial Networks
Jan 11, 2018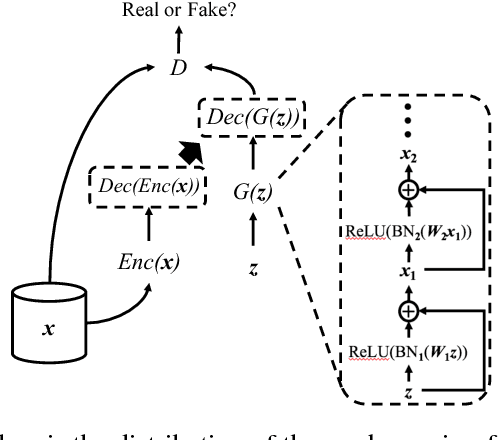
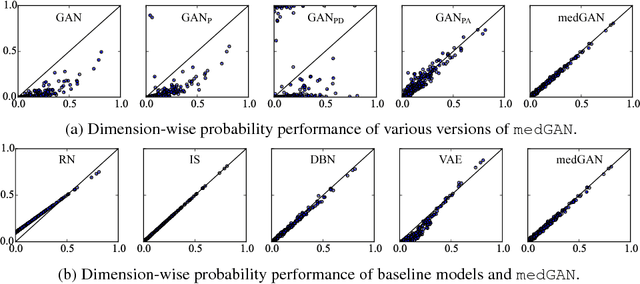
Abstract:Access to electronic health record (EHR) data has motivated computational advances in medical research. However, various concerns, particularly over privacy, can limit access to and collaborative use of EHR data. Sharing synthetic EHR data could mitigate risk. In this paper, we propose a new approach, medical Generative Adversarial Network (medGAN), to generate realistic synthetic patient records. Based on input real patient records, medGAN can generate high-dimensional discrete variables (e.g., binary and count features) via a combination of an autoencoder and generative adversarial networks. We also propose minibatch averaging to efficiently avoid mode collapse, and increase the learning efficiency with batch normalization and shortcut connections. To demonstrate feasibility, we showed that medGAN generates synthetic patient records that achieve comparable performance to real data on many experiments including distribution statistics, predictive modeling tasks and a medical expert review. We also empirically observe a limited privacy risk in both identity and attribute disclosure using medGAN.
SLEEPNET: Automated Sleep Staging System via Deep Learning
Jul 26, 2017



Abstract:Sleep disorders, such as sleep apnea, parasomnias, and hypersomnia, affect 50-70 million adults in the United States (Hillman et al., 2006). Overnight polysomnography (PSG), including brain monitoring using electroencephalography (EEG), is a central component of the diagnostic evaluation for sleep disorders. While PSG is conventionally performed by trained technologists, the recent rise of powerful neural network learning algorithms combined with large physiological datasets offers the possibility of automation, potentially making expert-level sleep analysis more widely available. We propose SLEEPNET (Sleep EEG neural network), a deployed annotation tool for sleep staging. SLEEPNET uses a deep recurrent neural network trained on the largest sleep physiology database assembled to date, consisting of PSGs from over 10,000 patients from the Massachusetts General Hospital (MGH) Sleep Laboratory. SLEEPNET achieves human-level annotation performance on an independent test set of 1,000 EEGs, with an average accuracy of 85.76% and algorithm-expert inter-rater agreement (IRA) of kappa = 79.46%, comparable to expert-expert IRA.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge