Ayis Pyrros
Shifts in Doctors' Eye Movements Between Real and AI-Generated Medical Images
Apr 21, 2025Abstract:Eye-tracking analysis plays a vital role in medical imaging, providing key insights into how radiologists visually interpret and diagnose clinical cases. In this work, we first analyze radiologists' attention and agreement by measuring the distribution of various eye-movement patterns, including saccades direction, amplitude, and their joint distribution. These metrics help uncover patterns in attention allocation and diagnostic strategies. Furthermore, we investigate whether and how doctors' gaze behavior shifts when viewing authentic (Real) versus deep-learning-generated (Fake) images. To achieve this, we examine fixation bias maps, focusing on first, last, short, and longest fixations independently, along with detailed saccades patterns, to quantify differences in gaze distribution and visual saliency between authentic and synthetic images.
Eyes Tell the Truth: GazeVal Highlights Shortcomings of Generative AI in Medical Imaging
Mar 26, 2025



Abstract:The demand for high-quality synthetic data for model training and augmentation has never been greater in medical imaging. However, current evaluations predominantly rely on computational metrics that fail to align with human expert recognition. This leads to synthetic images that may appear realistic numerically but lack clinical authenticity, posing significant challenges in ensuring the reliability and effectiveness of AI-driven medical tools. To address this gap, we introduce GazeVal, a practical framework that synergizes expert eye-tracking data with direct radiological evaluations to assess the quality of synthetic medical images. GazeVal leverages gaze patterns of radiologists as they provide a deeper understanding of how experts perceive and interact with synthetic data in different tasks (i.e., diagnostic or Turing tests). Experiments with sixteen radiologists revealed that 96.6% of the generated images (by the most recent state-of-the-art AI algorithm) were identified as fake, demonstrating the limitations of generative AI in producing clinically accurate images.
Large-Scale Multi-Center CT and MRI Segmentation of Pancreas with Deep Learning
May 20, 2024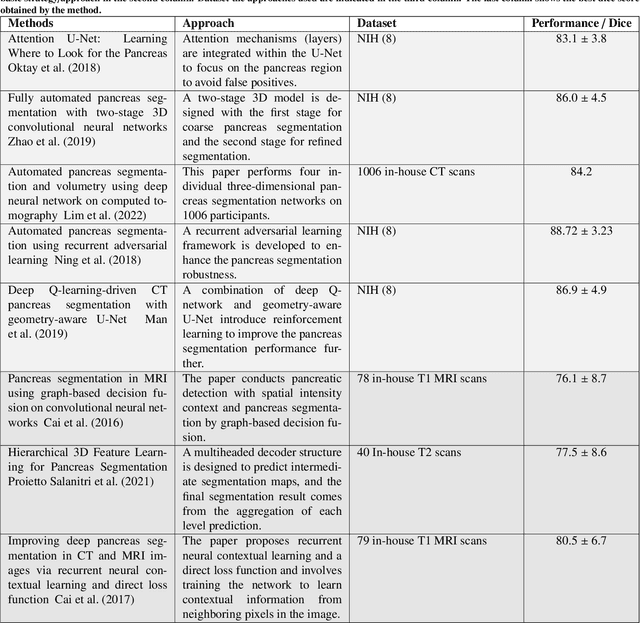
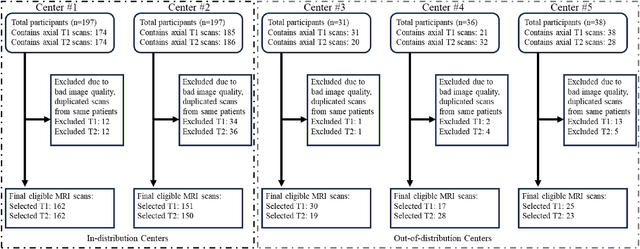
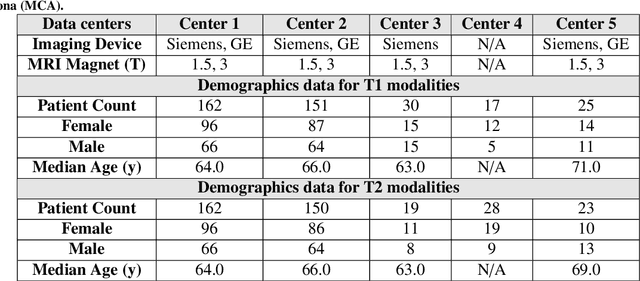
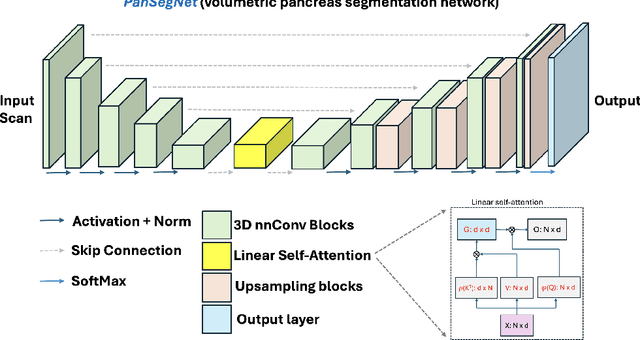
Abstract:Automated volumetric segmentation of the pancreas on cross-sectional imaging is needed for diagnosis and follow-up of pancreatic diseases. While CT-based pancreatic segmentation is more established, MRI-based segmentation methods are understudied, largely due to a lack of publicly available datasets, benchmarking research efforts, and domain-specific deep learning methods. In this retrospective study, we collected a large dataset (767 scans from 499 participants) of T1-weighted (T1W) and T2-weighted (T2W) abdominal MRI series from five centers between March 2004 and November 2022. We also collected CT scans of 1,350 patients from publicly available sources for benchmarking purposes. We developed a new pancreas segmentation method, called PanSegNet, combining the strengths of nnUNet and a Transformer network with a new linear attention module enabling volumetric computation. We tested PanSegNet's accuracy in cross-modality (a total of 2,117 scans) and cross-center settings with Dice and Hausdorff distance (HD95) evaluation metrics. We used Cohen's kappa statistics for intra and inter-rater agreement evaluation and paired t-tests for volume and Dice comparisons, respectively. For segmentation accuracy, we achieved Dice coefficients of 88.3% (std: 7.2%, at case level) with CT, 85.0% (std: 7.9%) with T1W MRI, and 86.3% (std: 6.4%) with T2W MRI. There was a high correlation for pancreas volume prediction with R^2 of 0.91, 0.84, and 0.85 for CT, T1W, and T2W, respectively. We found moderate inter-observer (0.624 and 0.638 for T1W and T2W MRI, respectively) and high intra-observer agreement scores. All MRI data is made available at https://osf.io/kysnj/. Our source code is available at https://github.com/NUBagciLab/PaNSegNet.
Reading Race: AI Recognises Patient's Racial Identity In Medical Images
Jul 21, 2021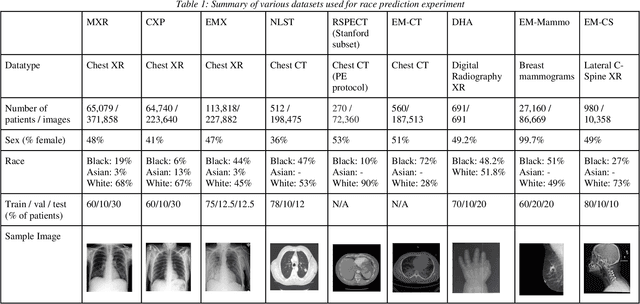
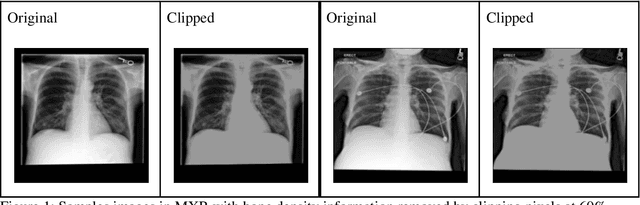
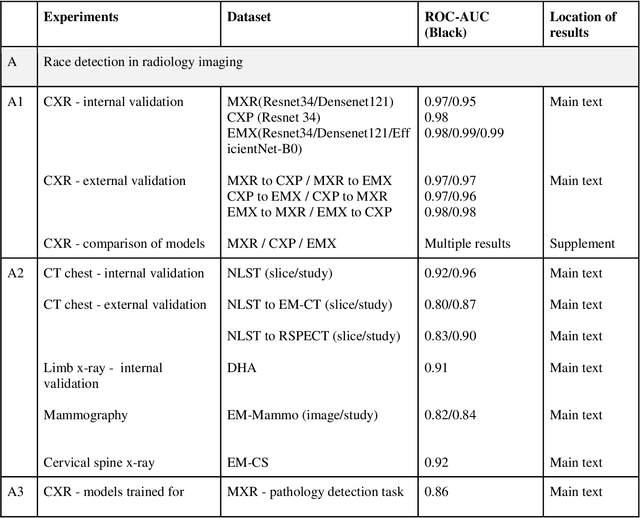
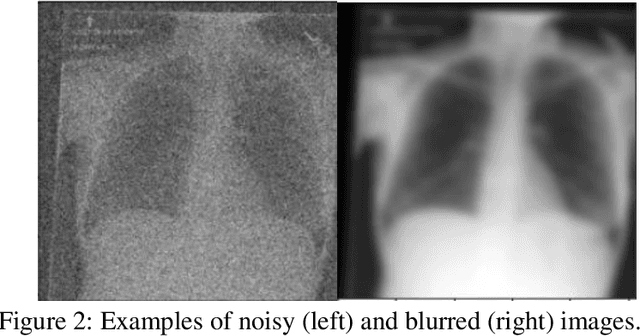
Abstract:Background: In medical imaging, prior studies have demonstrated disparate AI performance by race, yet there is no known correlation for race on medical imaging that would be obvious to the human expert interpreting the images. Methods: Using private and public datasets we evaluate: A) performance quantification of deep learning models to detect race from medical images, including the ability of these models to generalize to external environments and across multiple imaging modalities, B) assessment of possible confounding anatomic and phenotype population features, such as disease distribution and body habitus as predictors of race, and C) investigation into the underlying mechanism by which AI models can recognize race. Findings: Standard deep learning models can be trained to predict race from medical images with high performance across multiple imaging modalities. Our findings hold under external validation conditions, as well as when models are optimized to perform clinically motivated tasks. We demonstrate this detection is not due to trivial proxies or imaging-related surrogate covariates for race, such as underlying disease distribution. Finally, we show that performance persists over all anatomical regions and frequency spectrum of the images suggesting that mitigation efforts will be challenging and demand further study. Interpretation: We emphasize that model ability to predict self-reported race is itself not the issue of importance. However, our findings that AI can trivially predict self-reported race -- even from corrupted, cropped, and noised medical images -- in a setting where clinical experts cannot, creates an enormous risk for all model deployments in medical imaging: if an AI model secretly used its knowledge of self-reported race to misclassify all Black patients, radiologists would not be able to tell using the same data the model has access to.
EMIXER: End-to-end Multimodal X-ray Generation via Self-supervision
Jul 10, 2020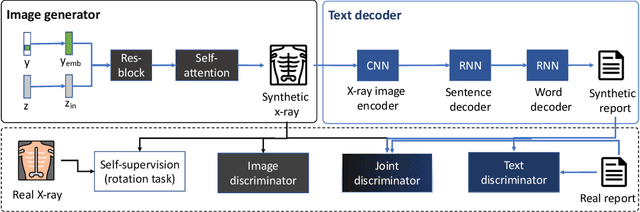

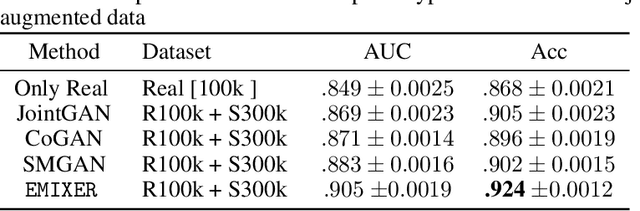

Abstract:Deep generative models have enabled the automated synthesis of high-quality data for diverse applications. However, the most effective generative models are specialized to data from a single domain (e.g., images or text). Real-world applications such as healthcare require multi-modal data from multiple domains (e.g., both images and corresponding text), which are difficult to acquire due to limited availability and privacy concerns and are much harder to synthesize. To tackle this joint synthesis challenge, we propose an End-to-end MultImodal X-ray genERative model (EMIXER) for jointly synthesizing x-ray images and corresponding free-text reports, all conditional on diagnosis labels. EMIXER is an conditional generative adversarial model by 1) generating an image based on a label, 2) encoding the image to a hidden embedding, 3) producing the corresponding text via a hierarchical decoder from the image embedding, and 4) a joint discriminator for assessing both the image and the corresponding text. EMIXER also enables self-supervision to leverage vast amount of unlabeled data. Extensive experiments with real X-ray reports data illustrate how data augmentation using synthesized multimodal samples can improve the performance of a variety of supervised tasks including COVID-19 X-ray classification with very limited samples. The quality of generated images and reports are also confirmed by radiologists. We quantitatively show that EMIXER generated synthetic datasets can augment X-ray image classification, report generation models to achieve 5.94% and 6.9% improvement on models trained only on real data samples. Taken together, our results highlight the promise of state of generative models to advance clinical machine learning.
Max-Sliced Wasserstein Distance and its use for GANs
Apr 11, 2019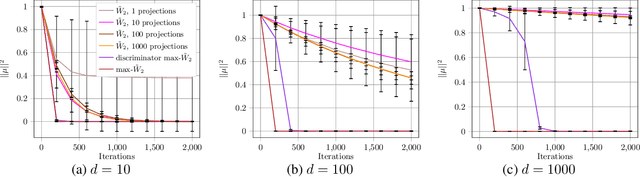
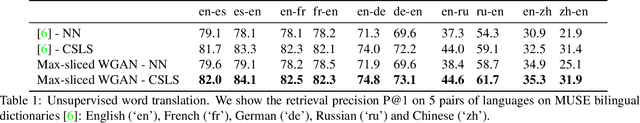
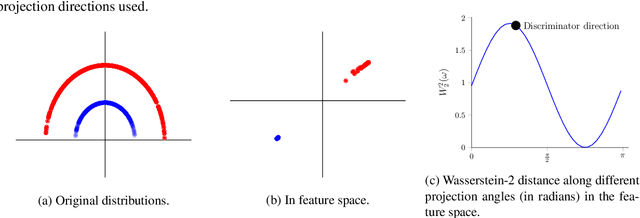
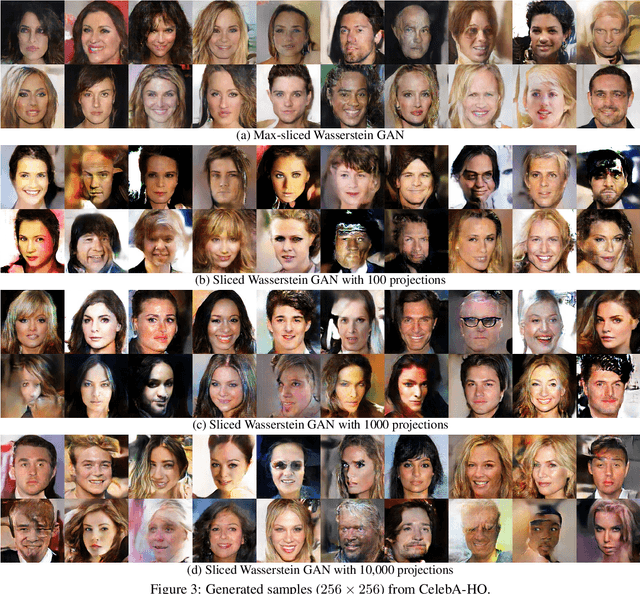
Abstract:Generative adversarial nets (GANs) and variational auto-encoders have significantly improved our distribution modeling capabilities, showing promise for dataset augmentation, image-to-image translation and feature learning. However, to model high-dimensional distributions, sequential training and stacked architectures are common, increasing the number of tunable hyper-parameters as well as the training time. Nonetheless, the sample complexity of the distance metrics remains one of the factors affecting GAN training. We first show that the recently proposed sliced Wasserstein distance has compelling sample complexity properties when compared to the Wasserstein distance. To further improve the sliced Wasserstein distance we then analyze its `projection complexity' and develop the max-sliced Wasserstein distance which enjoys compelling sample complexity while reducing projection complexity, albeit necessitating a max estimation. We finally illustrate that the proposed distance trains GANs on high-dimensional images up to a resolution of 256x256 easily.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge