M Brandon Westover
ManyDG: Many-domain Generalization for Healthcare Applications
Jan 21, 2023Abstract:The vast amount of health data has been continuously collected for each patient, providing opportunities to support diverse healthcare predictive tasks such as seizure detection and hospitalization prediction. Existing models are mostly trained on other patients data and evaluated on new patients. Many of them might suffer from poor generalizability. One key reason can be overfitting due to the unique information related to patient identities and their data collection environments, referred to as patient covariates in the paper. These patient covariates usually do not contribute to predicting the targets but are often difficult to remove. As a result, they can bias the model training process and impede generalization. In healthcare applications, most existing domain generalization methods assume a small number of domains. In this paper, considering the diversity of patient covariates, we propose a new setting by treating each patient as a separate domain (leading to many domains). We develop a new domain generalization method ManyDG, that can scale to such many-domain problems. Our method identifies the patient domain covariates by mutual reconstruction and removes them via an orthogonal projection step. Extensive experiments show that ManyDG can boost the generalization performance on multiple real-world healthcare tasks (e.g., 3.7% Jaccard improvements on MIMIC drug recommendation) and support realistic but challenging settings such as insufficient data and continuous learning.
Augmented Tensor Decomposition with Stochastic Optimization
Jul 14, 2021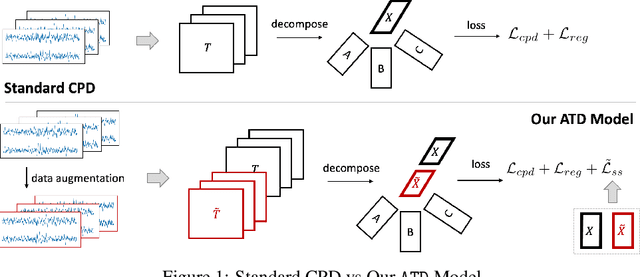
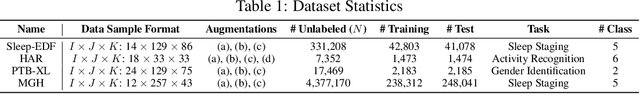
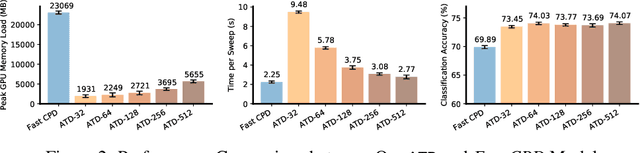
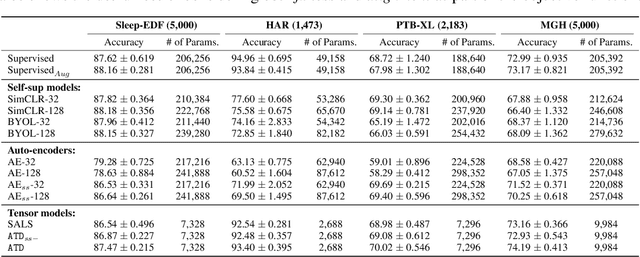
Abstract:Tensor decompositions are powerful tools for dimensionality reduction and feature interpretation of multidimensional data such as signals. Existing tensor decomposition objectives (e.g., Frobenius norm) are designed for fitting raw data under statistical assumptions, which may not align with downstream classification tasks. Also, real-world tensor data are usually high-ordered and have large dimensions with millions or billions of entries. Thus, it is expensive to decompose the whole tensor with traditional algorithms. In practice, raw tensor data also contains redundant information while data augmentation techniques may be used to smooth out noise in samples. This paper addresses the above challenges by proposing augmented tensor decomposition (ATD), which effectively incorporates data augmentations to boost downstream classification. To reduce the memory footprint of the decomposition, we propose a stochastic algorithm that updates the factor matrices in a batch fashion. We evaluate ATD on multiple signal datasets. It shows comparable or better performance (e.g., up to 15% in accuracy) over self-supervised and autoencoder baselines with less than 5% of model parameters, achieves 0.6% ~ 1.3% accuracy gain over other tensor-based baselines, and reduces the memory footprint by 9X when compared to standard tensor decomposition algorithms.
Automated Respiratory Event Detection Using Deep Neural Networks
Jan 12, 2021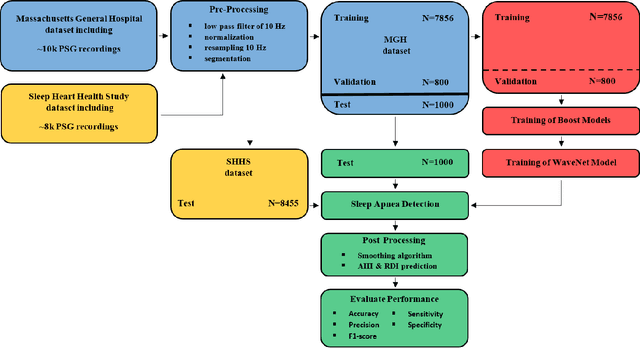
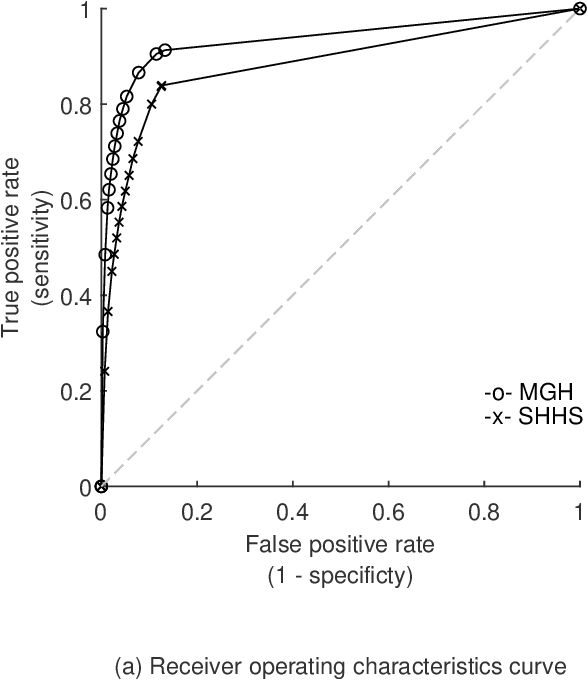
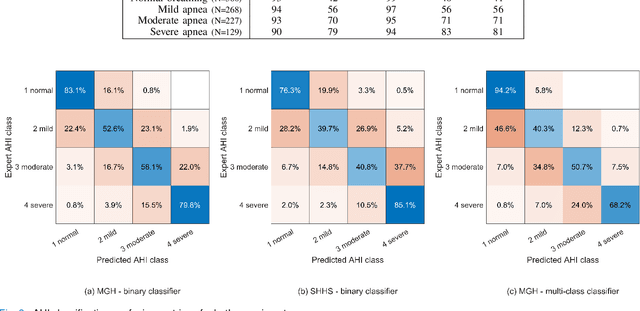
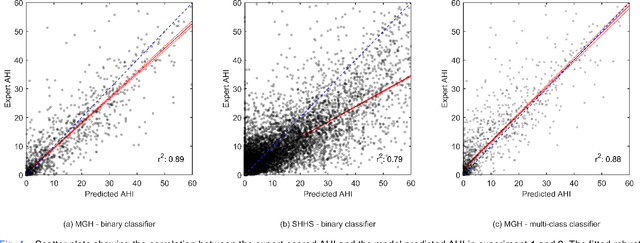
Abstract:The gold standard to assess respiration during sleep is polysomnography; a technique that is burdensome, expensive (both in analysis time and measurement costs), and difficult to repeat. Automation of respiratory analysis can improve test efficiency and enable accessible implementation opportunities worldwide. Using 9,656 polysomnography recordings from the Massachusetts General Hospital (MGH), we trained a neural network (WaveNet) based on a single respiratory effort belt to detect obstructive apnea, central apnea, hypopnea and respiratory-effort related arousals. Performance evaluation included event-based and recording-based metrics - using an apnea-hypopnea index analysis. The model was further evaluated on a public dataset, the Sleep-Heart-Health-Study-1, containing 8,455 polysomnographic recordings. For binary apnea event detection in the MGH dataset, the neural network obtained an accuracy of 95%, an apnea-hypopnea index $r^2$ of 0.89 and area under the curve for the receiver operating characteristics curve and precision-recall curve of 0.93 and 0.74, respectively. For the multiclass task, we obtained varying performances: 81% of all labeled central apneas were correctly classified, whereas this metric was 46% for obstructive apneas, 29% for respiratory effort related arousals and 16% for hypopneas. The majority of false predictions were misclassifications as another type of respiratory event. Our fully automated method can detect respiratory events and assess the apnea-hypopnea index with sufficient accuracy for clinical utilization. Differentiation of event types is more difficult and may reflect in part the complexity of human respiratory output and some degree of arbitrariness in the clinical thresholds and criteria used during manual annotation.
SLEEPNET: Automated Sleep Staging System via Deep Learning
Jul 26, 2017



Abstract:Sleep disorders, such as sleep apnea, parasomnias, and hypersomnia, affect 50-70 million adults in the United States (Hillman et al., 2006). Overnight polysomnography (PSG), including brain monitoring using electroencephalography (EEG), is a central component of the diagnostic evaluation for sleep disorders. While PSG is conventionally performed by trained technologists, the recent rise of powerful neural network learning algorithms combined with large physiological datasets offers the possibility of automation, potentially making expert-level sleep analysis more widely available. We propose SLEEPNET (Sleep EEG neural network), a deployed annotation tool for sleep staging. SLEEPNET uses a deep recurrent neural network trained on the largest sleep physiology database assembled to date, consisting of PSGs from over 10,000 patients from the Massachusetts General Hospital (MGH) Sleep Laboratory. SLEEPNET achieves human-level annotation performance on an independent test set of 1,000 EEGs, with an average accuracy of 85.76% and algorithm-expert inter-rater agreement (IRA) of kappa = 79.46%, comparable to expert-expert IRA.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge