Xiaorui Su
TxAgent: An AI Agent for Therapeutic Reasoning Across a Universe of Tools
Mar 14, 2025Abstract:Precision therapeutics require multimodal adaptive models that generate personalized treatment recommendations. We introduce TxAgent, an AI agent that leverages multi-step reasoning and real-time biomedical knowledge retrieval across a toolbox of 211 tools to analyze drug interactions, contraindications, and patient-specific treatment strategies. TxAgent evaluates how drugs interact at molecular, pharmacokinetic, and clinical levels, identifies contraindications based on patient comorbidities and concurrent medications, and tailors treatment strategies to individual patient characteristics. It retrieves and synthesizes evidence from multiple biomedical sources, assesses interactions between drugs and patient conditions, and refines treatment recommendations through iterative reasoning. It selects tools based on task objectives and executes structured function calls to solve therapeutic tasks that require clinical reasoning and cross-source validation. The ToolUniverse consolidates 211 tools from trusted sources, including all US FDA-approved drugs since 1939 and validated clinical insights from Open Targets. TxAgent outperforms leading LLMs, tool-use models, and reasoning agents across five new benchmarks: DrugPC, BrandPC, GenericPC, TreatmentPC, and DescriptionPC, covering 3,168 drug reasoning tasks and 456 personalized treatment scenarios. It achieves 92.1% accuracy in open-ended drug reasoning tasks, surpassing GPT-4o and outperforming DeepSeek-R1 (671B) in structured multi-step reasoning. TxAgent generalizes across drug name variants and descriptions. By integrating multi-step inference, real-time knowledge grounding, and tool-assisted decision-making, TxAgent ensures that treatment recommendations align with established clinical guidelines and real-world evidence, reducing the risk of adverse events and improving therapeutic decision-making.
Multimodal Medical Code Tokenizer
Feb 06, 2025



Abstract:Foundation models trained on patient electronic health records (EHRs) require tokenizing medical data into sequences of discrete vocabulary items. Existing tokenizers treat medical codes from EHRs as isolated textual tokens. However, each medical code is defined by its textual description, its position in ontological hierarchies, and its relationships to other codes, such as disease co-occurrences and drug-treatment associations. Medical vocabularies contain more than 600,000 codes with critical information for clinical reasoning. We introduce MedTok, a multimodal medical code tokenizer that uses the text descriptions and relational context of codes. MedTok processes text using a language model encoder and encodes the relational structure with a graph encoder. It then quantizes both modalities into a unified token space, preserving modality-specific and cross-modality information. We integrate MedTok into five EHR models and evaluate it on operational and clinical tasks across in-patient and out-patient datasets, including outcome prediction, diagnosis classification, drug recommendation, and risk stratification. Swapping standard EHR tokenizers with MedTok improves AUPRC across all EHR models, by 4.10% on MIMIC-III, 4.78% on MIMIC-IV, and 11.30% on EHRShot, with the largest gains in drug recommendation. Beyond EHR modeling, we demonstrate using MedTok tokenizer with medical QA systems. Our results demonstrate the potential of MedTok as a unified tokenizer for medical codes, improving tokenization for medical foundation models.
Knowledge Graph Based Agent for Complex, Knowledge-Intensive QA in Medicine
Oct 07, 2024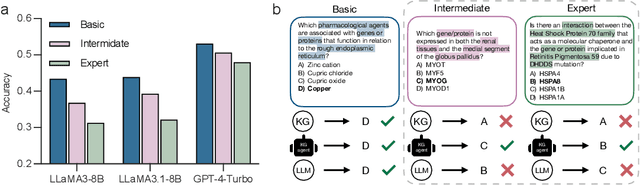
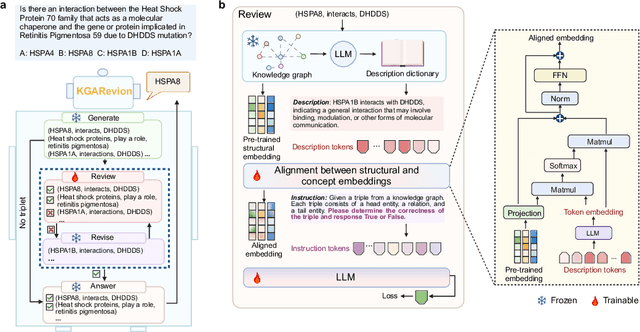
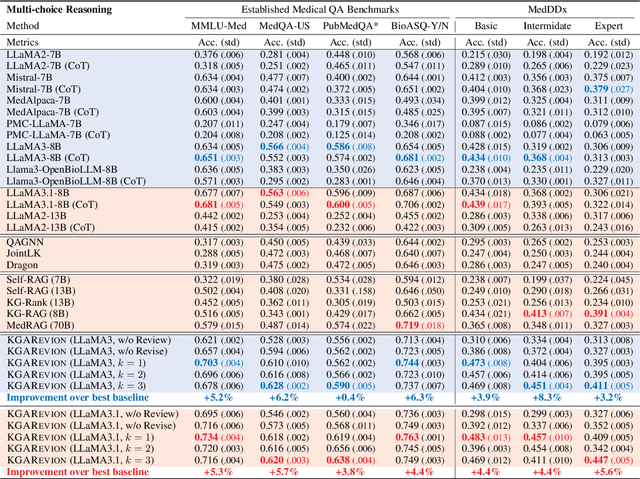
Abstract:Biomedical knowledge is uniquely complex and structured, requiring distinct reasoning strategies compared to other scientific disciplines like physics or chemistry. Biomedical scientists do not rely on a single approach to reasoning; instead, they use various strategies, including rule-based, prototype-based, and case-based reasoning. This diversity calls for flexible approaches that accommodate multiple reasoning strategies while leveraging in-domain knowledge. We introduce KGARevion, a knowledge graph (KG) based agent designed to address the complexity of knowledge-intensive medical queries. Upon receiving a query, KGARevion generates relevant triplets by using the knowledge base of the LLM. These triplets are then verified against a grounded KG to filter out erroneous information and ensure that only accurate, relevant data contribute to the final answer. Unlike RAG-based models, this multi-step process ensures robustness in reasoning while adapting to different models of medical reasoning. Evaluations on four gold-standard medical QA datasets show that KGARevion improves accuracy by over 5.2%, outperforming 15 models in handling complex medical questions. To test its capabilities, we curated three new medical QA datasets with varying levels of semantic complexity, where KGARevion achieved a 10.4% improvement in accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge