Valentina Giunchiglia
Application of Whisper in Clinical Practice: the Post-Stroke Speech Assessment during a Naming Task
Jul 23, 2025Abstract:Detailed assessment of language impairment following stroke remains a cognitively complex and clinician-intensive task, limiting timely and scalable diagnosis. Automatic Speech Recognition (ASR) foundation models offer a promising pathway to augment human evaluation through intelligent systems, but their effectiveness in the context of speech and language impairment remains uncertain. In this study, we evaluate whether Whisper, a state-of-the-art ASR foundation model, can be applied to transcribe and analyze speech from patients with stroke during a commonly used picture-naming task. We assess both verbatim transcription accuracy and the model's ability to support downstream prediction of language function, which has major implications for outcomes after stroke. Our results show that the baseline Whisper model performs poorly on single-word speech utterances. Nevertheless, fine-tuning Whisper significantly improves transcription accuracy (reducing Word Error Rate by 87.72% in healthy speech and 71.22% in speech from patients). Further, learned representations from the model enable accurate prediction of speech quality (average F1 Macro of 0.74 for healthy, 0.75 for patients). However, evaluations on an unseen (TORGO) dataset reveal limited generalizability, highlighting the inability of Whisper to perform zero-shot transcription of single-word utterances on out-of-domain clinical speech and emphasizing the need to adapt models to specific clinical populations. While challenges remain in cross-domain generalization, these findings highlight the potential of foundation models, when appropriately fine-tuned, to advance automated speech and language assessment and rehabilitation for stroke-related impairments.
Knowledge Graph Based Agent for Complex, Knowledge-Intensive QA in Medicine
Oct 07, 2024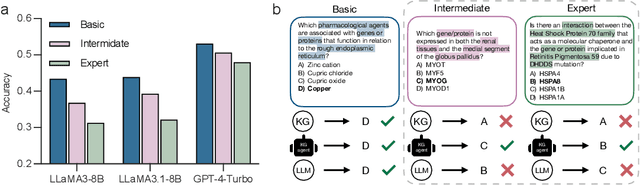
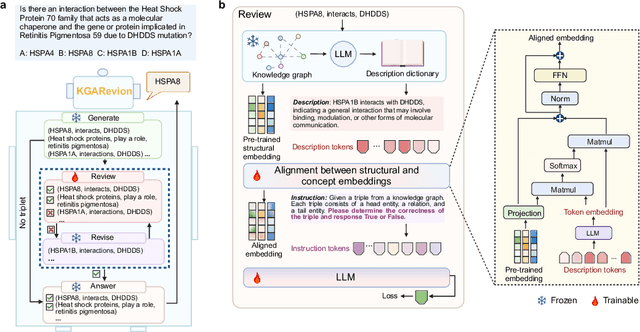
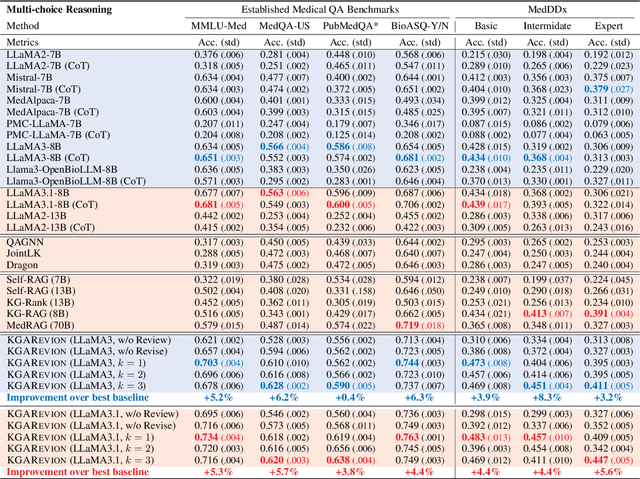
Abstract:Biomedical knowledge is uniquely complex and structured, requiring distinct reasoning strategies compared to other scientific disciplines like physics or chemistry. Biomedical scientists do not rely on a single approach to reasoning; instead, they use various strategies, including rule-based, prototype-based, and case-based reasoning. This diversity calls for flexible approaches that accommodate multiple reasoning strategies while leveraging in-domain knowledge. We introduce KGARevion, a knowledge graph (KG) based agent designed to address the complexity of knowledge-intensive medical queries. Upon receiving a query, KGARevion generates relevant triplets by using the knowledge base of the LLM. These triplets are then verified against a grounded KG to filter out erroneous information and ensure that only accurate, relevant data contribute to the final answer. Unlike RAG-based models, this multi-step process ensures robustness in reasoning while adapting to different models of medical reasoning. Evaluations on four gold-standard medical QA datasets show that KGARevion improves accuracy by over 5.2%, outperforming 15 models in handling complex medical questions. To test its capabilities, we curated three new medical QA datasets with varying levels of semantic complexity, where KGARevion achieved a 10.4% improvement in accuracy.
Empowering Biomedical Discovery with AI Agents
Apr 03, 2024



Abstract:We envision 'AI scientists' as systems capable of skeptical learning and reasoning that empower biomedical research through collaborative agents that integrate machine learning tools with experimental platforms. Rather than taking humans out of the discovery process, biomedical AI agents combine human creativity and expertise with AI's ability to analyze large datasets, navigate hypothesis spaces, and execute repetitive tasks. AI agents are proficient in a variety of tasks, including self-assessment and planning of discovery workflows. These agents use large language models and generative models to feature structured memory for continual learning and use machine learning tools to incorporate scientific knowledge, biological principles, and theories. AI agents can impact areas ranging from hybrid cell simulation, programmable control of phenotypes, and the design of cellular circuits to the development of new therapies.
Towards Training GNNs using Explanation Directed Message Passing
Dec 01, 2022Abstract:With the increasing use of Graph Neural Networks (GNNs) in critical real-world applications, several post hoc explanation methods have been proposed to understand their predictions. However, there has been no work in generating explanations on the fly during model training and utilizing them to improve the expressive power of the underlying GNN models. In this work, we introduce a novel explanation-directed neural message passing framework for GNNs, EXPASS (EXplainable message PASSing), which aggregates only embeddings from nodes and edges identified as important by a GNN explanation method. EXPASS can be used with any existing GNN architecture and subgraph-optimizing explainer to learn accurate graph embeddings. We theoretically show that EXPASS alleviates the oversmoothing problem in GNNs by slowing the layer wise loss of Dirichlet energy and that the embedding difference between the vanilla message passing and EXPASS framework can be upper bounded by the difference of their respective model weights. Our empirical results show that graph embeddings learned using EXPASS improve the predictive performance and alleviate the oversmoothing problems of GNNs, opening up new frontiers in graph machine learning to develop explanation-based training frameworks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge