Guadalupe Gonzalez
Towards Training GNNs using Explanation Directed Message Passing
Dec 01, 2022Abstract:With the increasing use of Graph Neural Networks (GNNs) in critical real-world applications, several post hoc explanation methods have been proposed to understand their predictions. However, there has been no work in generating explanations on the fly during model training and utilizing them to improve the expressive power of the underlying GNN models. In this work, we introduce a novel explanation-directed neural message passing framework for GNNs, EXPASS (EXplainable message PASSing), which aggregates only embeddings from nodes and edges identified as important by a GNN explanation method. EXPASS can be used with any existing GNN architecture and subgraph-optimizing explainer to learn accurate graph embeddings. We theoretically show that EXPASS alleviates the oversmoothing problem in GNNs by slowing the layer wise loss of Dirichlet energy and that the embedding difference between the vanilla message passing and EXPASS framework can be upper bounded by the difference of their respective model weights. Our empirical results show that graph embeddings learned using EXPASS improve the predictive performance and alleviate the oversmoothing problems of GNNs, opening up new frontiers in graph machine learning to develop explanation-based training frameworks.
Unsupervised Heterophilous Network Embedding via r-Ego Network Discrimination
Mar 28, 2022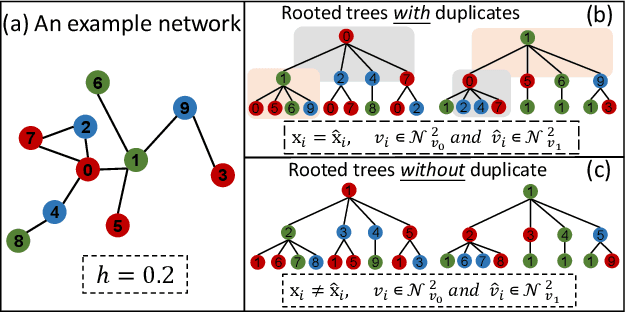
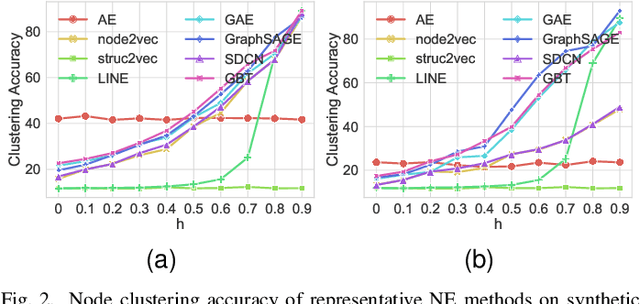
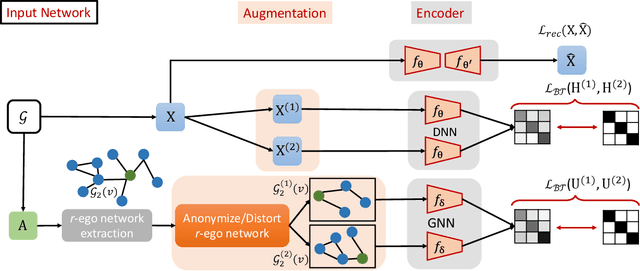
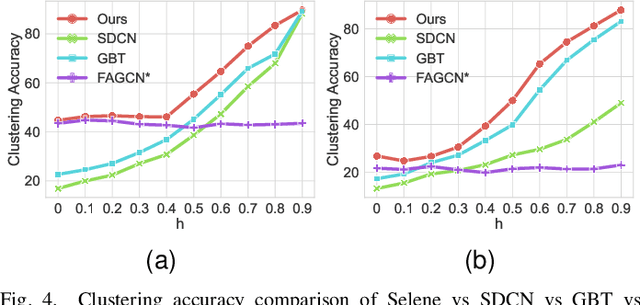
Abstract:Recently, supervised network embedding (NE) has emerged as a predominant technique for representing complex systems that take the form of networks, and various downstream node- and network-level tasks have benefited from its remarkable developments. However, unsupervised NE still remains challenging due to the uncertainty in defining a learning objective. In addition, it is still an unexplored research question whether existing NE methods adapt well to heterophilous networks. This paper introduces the first empirical study on the influence of homophily ratio on the performance of existing unsupervised NE methods and reveals their limitations. Inspired by our empirical findings, we design unsupervised NE task as an r-ego network discrimination problem and further develop a SELf-supErvised Network Embedding (Selene) framework for learning useful node representations for both homophilous and heterophilous networks. Specifically, we propose a dual-channel feature embedding mechanism to fuse node attributes and network structure information and leverage a sampling and anonymisation strategy to break the implicit homophily assumption of existing embedding mechanisms. Lastly, we introduce a negative-sample-free SSL objective function to optimise the framework. We conduct extensive experiments and a series of ablation studies on 12 real-world datasets and 20 synthetic networks. Results demonstrate Selene's superior performance and confirm the effectiveness of each component.
Graph Attentional Autoencoder for Anticancer Hyperfood Prediction
Jan 16, 2020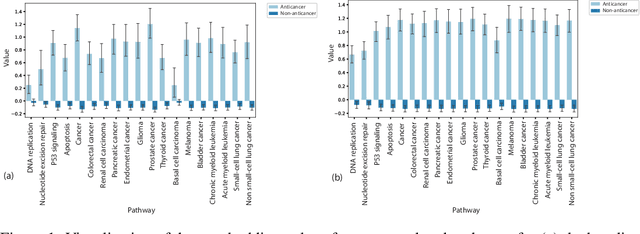
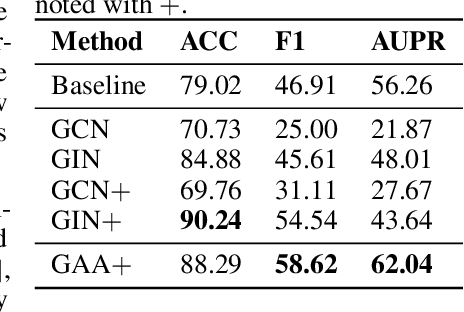
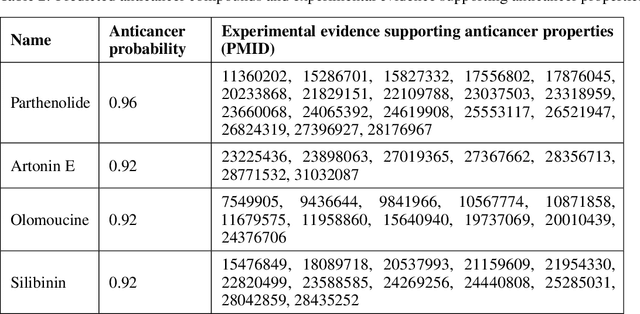
Abstract:Recent research efforts have shown the possibility to discover anticancer drug-like molecules in food from their effect on protein-protein interaction networks, opening a potential pathway to disease-beating diet design. We formulate this task as a graph classification problem on which graph neural networks (GNNs) have achieved state-of-the-art results. However, GNNs are difficult to train on sparse low-dimensional features according to our empirical evidence. Here, we present graph augmented features, integrating graph structural information and raw node attributes with varying ratios, to ease the training of networks. We further introduce a novel neural network architecture on graphs, the Graph Attentional Autoencoder (GAA) to predict food compounds with anticancer properties based on perturbed protein networks. We demonstrate that the method outperforms the baseline approach and state-of-the-art graph classification models in this task.
Learning Interpretable Disease Self-Representations for Drug Repositioning
Oct 20, 2019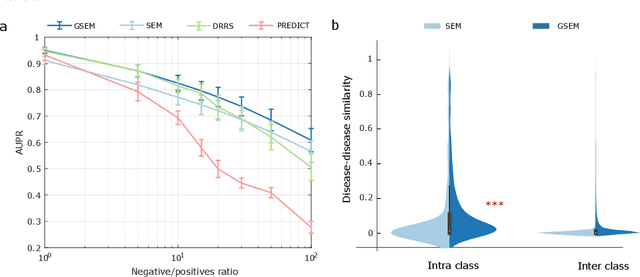

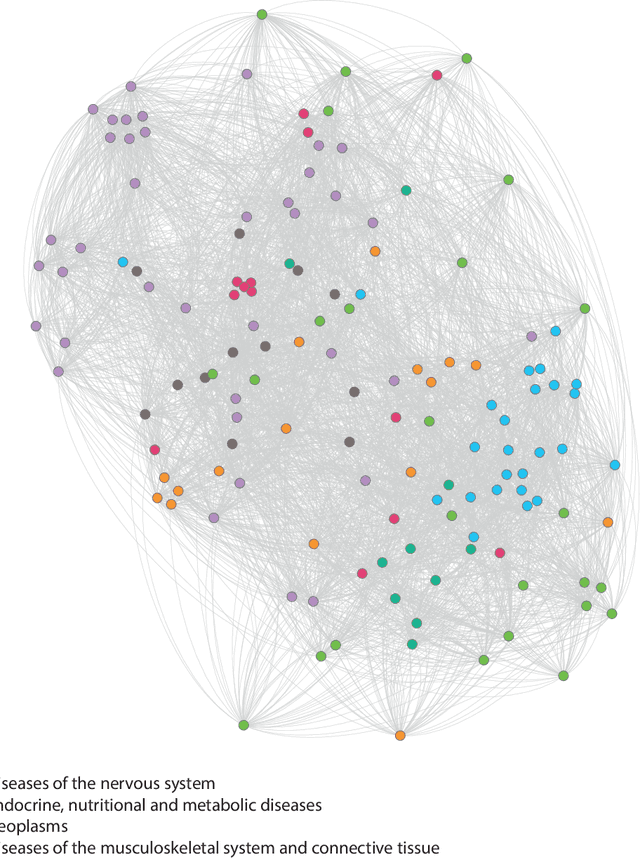
Abstract:Drug repositioning is an attractive cost-efficient strategy for the development of treatments for human diseases. Here, we propose an interpretable model that learns disease self-representations for drug repositioning. Our self-representation model represents each disease as a linear combination of a few other diseases. We enforce proximity in the learnt representations in a way to preserve the geometric structure of the human phenome network - a domain-specific knowledge that naturally adds relational inductive bias to the disease self-representations. We prove that our method is globally optimal and show results outperforming state-of-the-art drug repositioning approaches. We further show that the disease self-representations are biologically interpretable.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge