Ivan Laponogov
The Helicobacter pylori AI-Clinician: Harnessing Artificial Intelligence to Personalize H. pylori Treatment Recommendations
Dec 07, 2024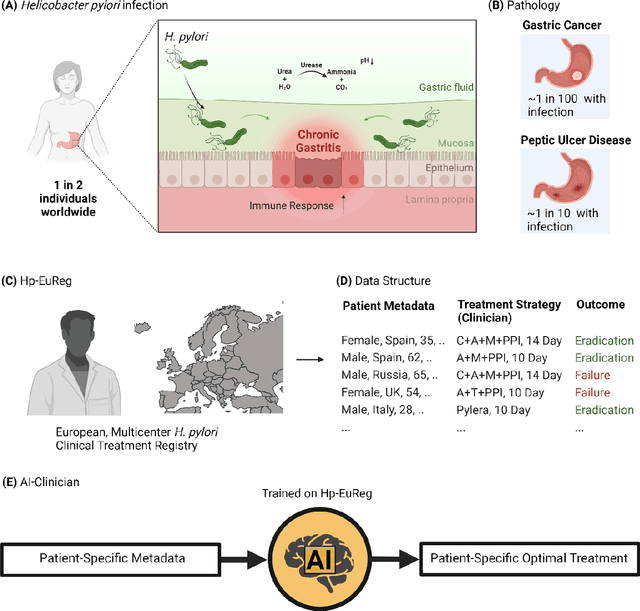
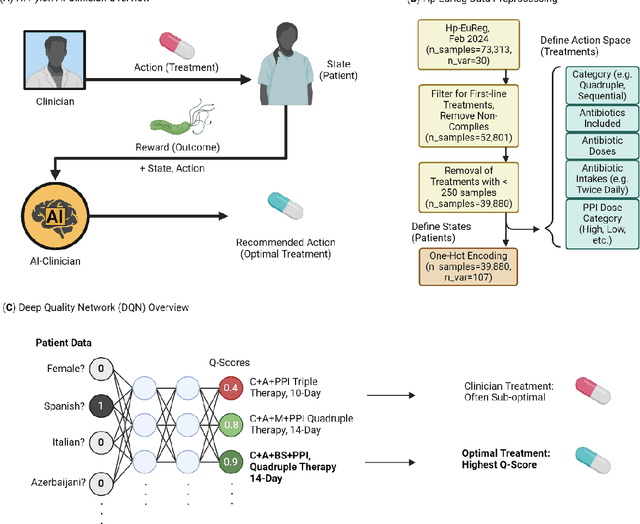
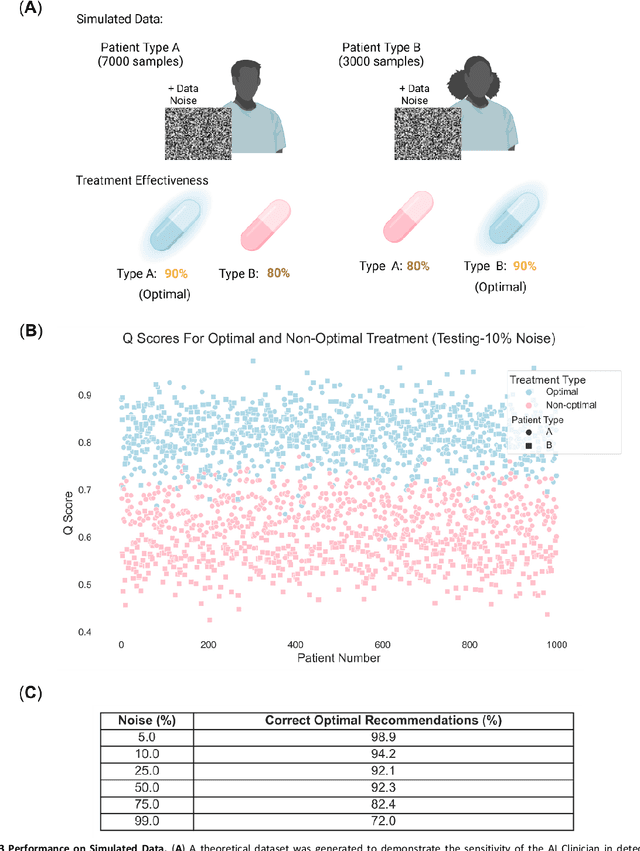
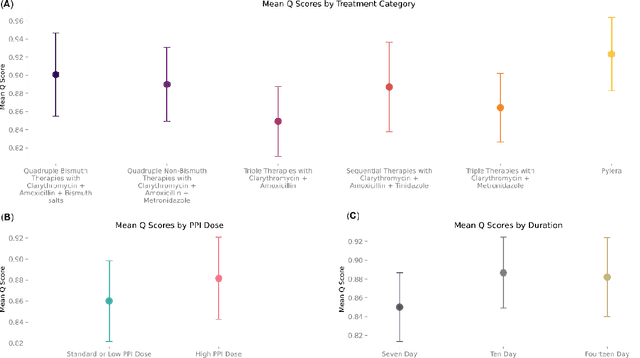
Abstract:Helicobacter pylori (H. pylori) is the most common carcinogenic pathogen worldwide. Infecting roughly 1 in 2 individuals globally, it is the leading cause of peptic ulcer disease, chronic gastritis, and gastric cancer. To investigate whether personalized treatments would be optimal for patients suffering from infection, we developed the H. pylori AI-clinician recommendation system. This system was trained on data from tens of thousands of H. pylori-infected patients from Hp-EuReg, orders of magnitude greater than those experienced by a single real-world clinician. We first used a simulated dataset and demonstrated the ability of our AI Clinician method to identify patient subgroups that would benefit from differential optimal treatments. Next, we trained the AI Clinician on Hp-EuReg, demonstrating the AI Clinician reproduces known quality estimates of treatments, for example bismuth and quadruple therapies out-performing triple, with longer durations and higher dose proton pump inhibitor (PPI) showing higher quality estimation on average. Next we demonstrated that treatment was optimized by recommended personalized therapies in patient subsets, where 65% of patients were recommended a bismuth therapy of either metronidazole, tetracycline, and bismuth salts with PPI, or bismuth quadruple therapy with clarithromycin, amoxicillin, and bismuth salts with PPI, and 15% of patients recommended a quadruple non-bismuth therapy of clarithromycin, amoxicillin, and metronidazole with PPI. Finally, we determined trends in patient variables driving the personalized recommendations using random forest modelling. With around half of the world likely to experience H. pylori infection at some point in their lives, the identification of personalized optimal treatments will be crucial in both gastric cancer prevention and quality of life improvements for countless individuals worldwide.
Optimizing Ingredient Substitution Using Large Language Models to Enhance Phytochemical Content in Recipes
Sep 13, 2024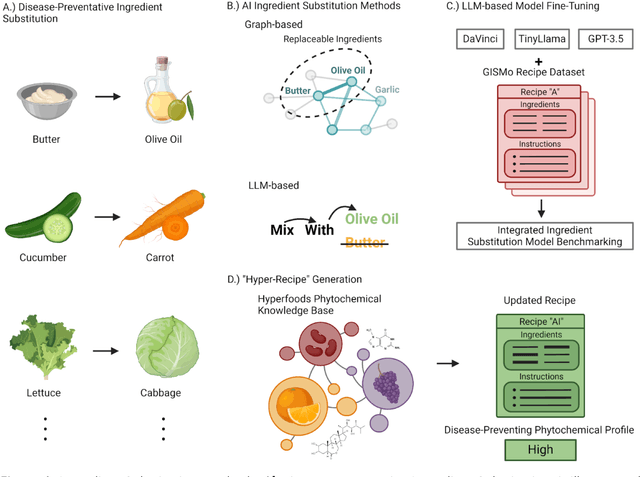
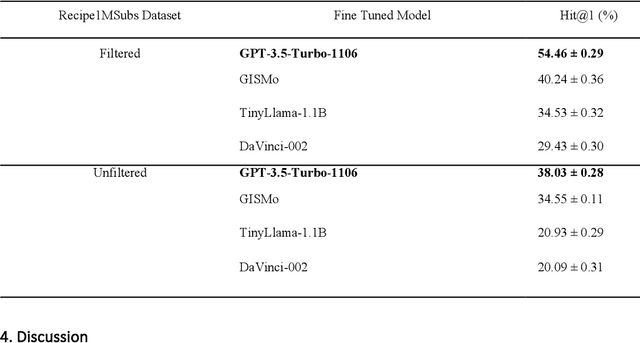
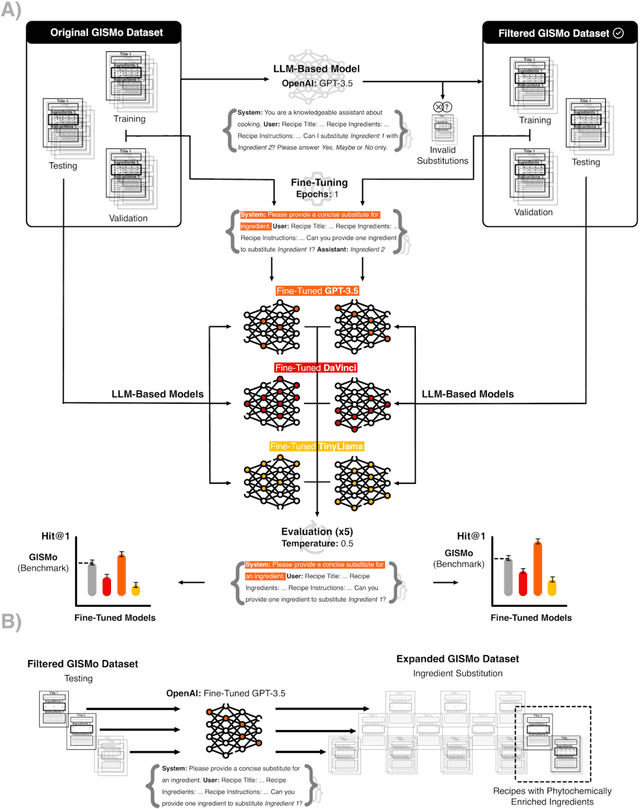
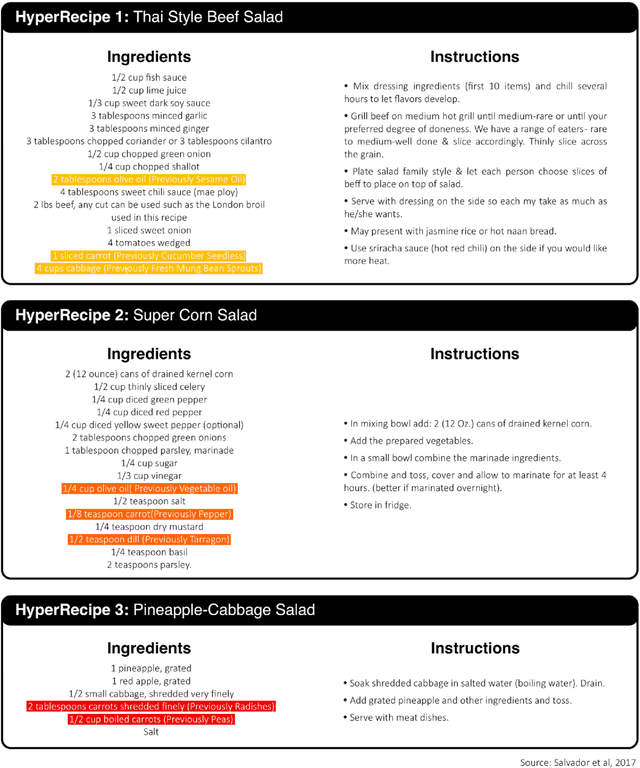
Abstract:In the emerging field of computational gastronomy, aligning culinary practices with scientifically supported nutritional goals is increasingly important. This study explores how large language models (LLMs) can be applied to optimize ingredient substitutions in recipes, specifically to enhance the phytochemical content of meals. Phytochemicals are bioactive compounds found in plants, which, based on preclinical studies, may offer potential health benefits. We fine-tuned models, including OpenAI's GPT-3.5, DaVinci, and Meta's TinyLlama, using an ingredient substitution dataset. These models were used to predict substitutions that enhance phytochemical content and create a corresponding enriched recipe dataset. Our approach improved Hit@1 accuracy on ingredient substitution tasks, from the baseline 34.53 plus-minus 0.10% to 38.03 plus-minus 0.28% on the original GISMo dataset, and from 40.24 plus-minus 0.36% to 54.46 plus-minus 0.29% on a refined version of the same dataset. These substitutions led to the creation of 1,951 phytochemically enriched ingredient pairings and 1,639 unique recipes. While this approach demonstrates potential in optimizing ingredient substitutions, caution must be taken when drawing conclusions about health benefits, as the claims are based on preclinical evidence. Future work should include clinical validation and broader datasets to further evaluate the nutritional impact of these substitutions. This research represents a step forward in using AI to promote healthier eating practices, providing potential pathways for integrating computational methods with nutritional science.
Foundational Models for Pathology and Endoscopy Images: Application for Gastric Inflammation
Jun 26, 2024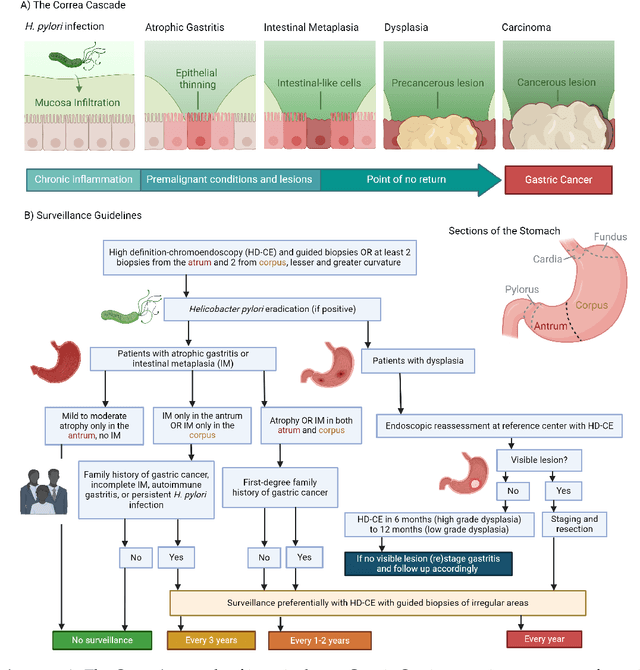
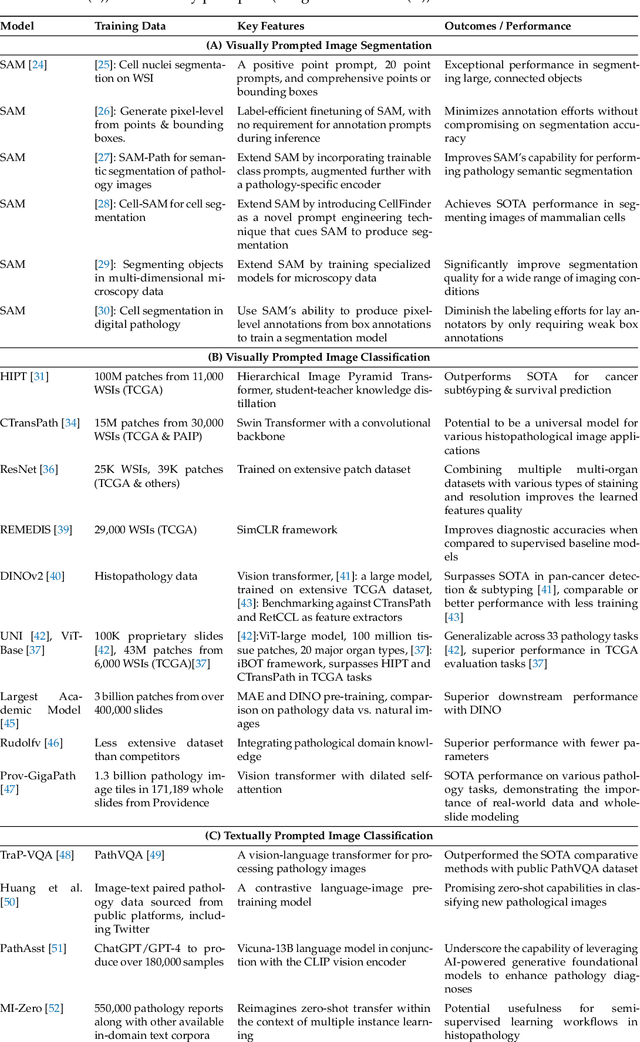
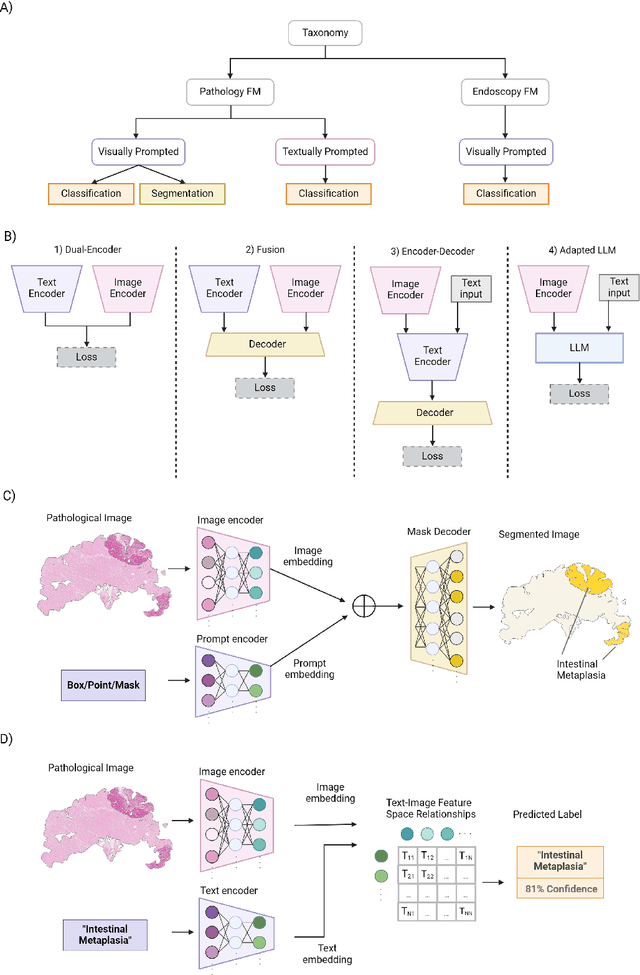
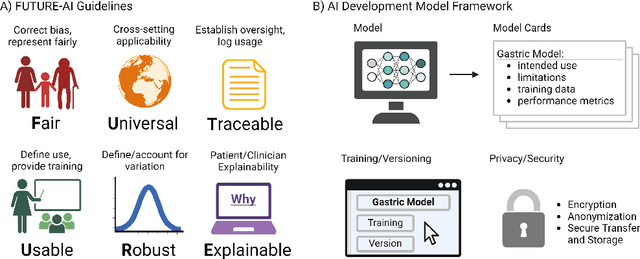
Abstract:The integration of artificial intelligence (AI) in medical diagnostics represents a significant advancement in managing upper gastrointestinal (GI) cancer, a major cause of global cancer mortality. Specifically for gastric cancer (GC), chronic inflammation causes changes in the mucosa such as atrophy, intestinal metaplasia (IM), dysplasia and ultimately cancer. Early detection through endoscopic regular surveillance is essential for better outcomes. Foundation models (FM), which are machine or deep learning models trained on diverse data and applicable to broad use cases, offer a promising solution to enhance the accuracy of endoscopy and its subsequent pathology image analysis. This review explores the recent advancements, applications, and challenges associated with FM in endoscopy and pathology imaging. We started by elucidating the core principles and architectures underlying these models, including their training methodologies and the pivotal role of large-scale data in developing their predictive capabilities. Moreover, this work discusses emerging trends and future research directions, emphasizing the integration of multimodal data, the development of more robust and equitable models, and the potential for real-time diagnostic support. This review aims to provide a roadmap for researchers and practitioners in navigating the complexities of incorporating FM into clinical practice for prevention/management of GC cases, thereby improving patient outcomes.
Graph Attentional Autoencoder for Anticancer Hyperfood Prediction
Jan 16, 2020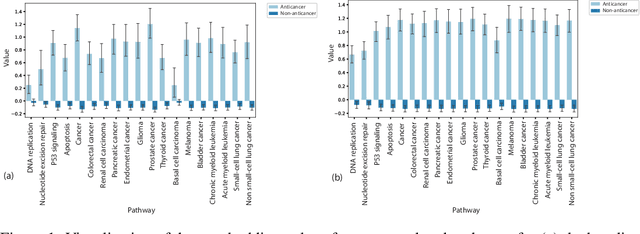
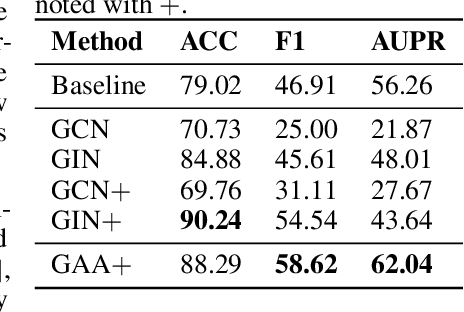
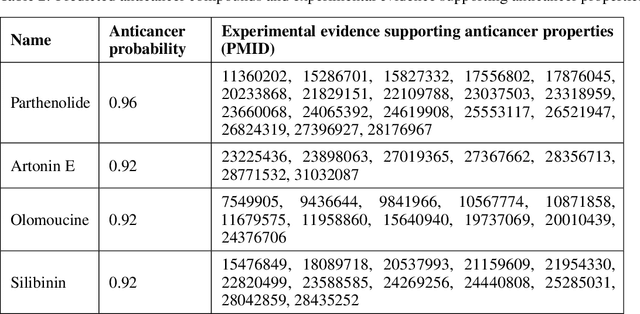
Abstract:Recent research efforts have shown the possibility to discover anticancer drug-like molecules in food from their effect on protein-protein interaction networks, opening a potential pathway to disease-beating diet design. We formulate this task as a graph classification problem on which graph neural networks (GNNs) have achieved state-of-the-art results. However, GNNs are difficult to train on sparse low-dimensional features according to our empirical evidence. Here, we present graph augmented features, integrating graph structural information and raw node attributes with varying ratios, to ease the training of networks. We further introduce a novel neural network architecture on graphs, the Graph Attentional Autoencoder (GAA) to predict food compounds with anticancer properties based on perturbed protein networks. We demonstrate that the method outperforms the baseline approach and state-of-the-art graph classification models in this task.
Learning Interpretable Disease Self-Representations for Drug Repositioning
Oct 20, 2019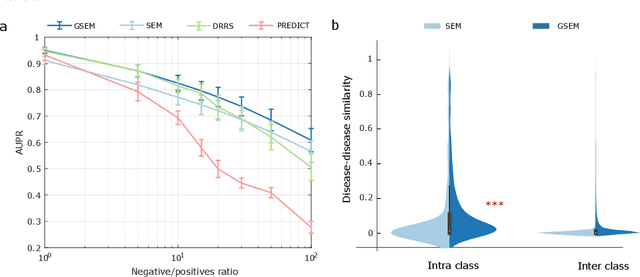

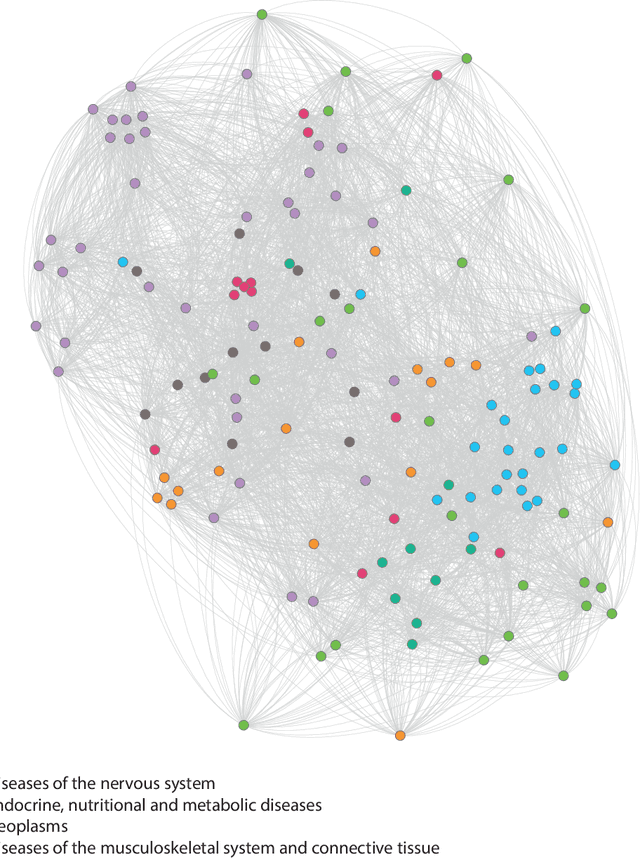
Abstract:Drug repositioning is an attractive cost-efficient strategy for the development of treatments for human diseases. Here, we propose an interpretable model that learns disease self-representations for drug repositioning. Our self-representation model represents each disease as a linear combination of a few other diseases. We enforce proximity in the learnt representations in a way to preserve the geometric structure of the human phenome network - a domain-specific knowledge that naturally adds relational inductive bias to the disease self-representations. We prove that our method is globally optimal and show results outperforming state-of-the-art drug repositioning approaches. We further show that the disease self-representations are biologically interpretable.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge