Owen Queen
DSGym: A Holistic Framework for Evaluating and Training Data Science Agents
Jan 22, 2026Abstract:Data science agents promise to accelerate discovery and insight-generation by turning data into executable analyses and findings. Yet existing data science benchmarks fall short due to fragmented evaluation interfaces that make cross-benchmark comparison difficult, narrow task coverage and a lack of rigorous data grounding. In particular, we show that a substantial portion of tasks in current benchmarks can be solved without using the actual data. To address these limitations, we introduce DSGym, a standardized framework for evaluating and training data science agents in self-contained execution environments. Unlike static benchmarks, DSGym provides a modular architecture that makes it easy to add tasks, agent scaffolds, and tools, positioning it as a live, extensible testbed. We curate DSGym-Tasks, a holistic task suite that standardizes and refines existing benchmarks via quality and shortcut solvability filtering. We further expand coverage with (1) DSBio: expert-derived bioinformatics tasks grounded in literature and (2) DSPredict: challenging prediction tasks spanning domains such as computer vision, molecular prediction, and single-cell perturbation. Beyond evaluation, DSGym enables agent training via execution-verified data synthesis pipeline. As a case study, we build a 2,000-example training set and trained a 4B model in DSGym that outperforms GPT-4o on standardized analysis benchmarks. Overall, DSGym enables rigorous end-to-end measurement of whether agents can plan, implement, and validate data analyses in realistic scientific context.
Exploring the use of AI authors and reviewers at Agents4Science
Nov 19, 2025Abstract:There is growing interest in using AI agents for scientific research, yet fundamental questions remain about their capabilities as scientists and reviewers. To explore these questions, we organized Agents4Science, the first conference in which AI agents serve as both primary authors and reviewers, with humans as co-authors and co-reviewers. Here, we discuss the key learnings from the conference and their implications for human-AI collaboration in science.
UniTS: Building a Unified Time Series Model
Feb 29, 2024



Abstract:Foundation models, especially LLMs, are profoundly transforming deep learning. Instead of training many task-specific models, we can adapt a single pretrained model to many tasks via fewshot prompting or fine-tuning. However, current foundation models apply to sequence data but not to time series, which present unique challenges due to the inherent diverse and multidomain time series datasets, diverging task specifications across forecasting, classification and other types of tasks, and the apparent need for task-specialized models. We developed UNITS, a unified time series model that supports a universal task specification, accommodating classification, forecasting, imputation, and anomaly detection tasks. This is achieved through a novel unified network backbone, which incorporates sequence and variable attention along with a dynamic linear operator and is trained as a unified model. Across 38 multi-domain datasets, UNITS demonstrates superior performance compared to task-specific models and repurposed natural language-based LLMs. UNITS exhibits remarkable zero-shot, few-shot, and prompt learning capabilities when evaluated on new data domains and tasks. The source code and datasets are available at https://github.com/mims-harvard/UniTS.
Graph AI in Medicine
Oct 20, 2023Abstract:In clinical artificial intelligence (AI), graph representation learning, mainly through graph neural networks (GNNs), stands out for its capability to capture intricate relationships within structured clinical datasets. With diverse data -- from patient records to imaging -- GNNs process data holistically by viewing modalities as nodes interconnected by their relationships. Graph AI facilitates model transfer across clinical tasks, enabling models to generalize across patient populations without additional parameters or minimal re-training. However, the importance of human-centered design and model interpretability in clinical decision-making cannot be overstated. Since graph AI models capture information through localized neural transformations defined on graph relationships, they offer both an opportunity and a challenge in elucidating model rationale. Knowledge graphs can enhance interpretability by aligning model-driven insights with medical knowledge. Emerging graph models integrate diverse data modalities through pre-training, facilitate interactive feedback loops, and foster human-AI collaboration, paving the way to clinically meaningful predictions.
Encoding Time-Series Explanations through Self-Supervised Model Behavior Consistency
Jun 03, 2023Abstract:Interpreting time series models is uniquely challenging because it requires identifying both the location of time series signals that drive model predictions and their matching to an interpretable temporal pattern. While explainers from other modalities can be applied to time series, their inductive biases do not transfer well to the inherently uninterpretable nature of time series. We present TimeX, a time series consistency model for training explainers. TimeX trains an interpretable surrogate to mimic the behavior of a pretrained time series model. It addresses the issue of model faithfulness by introducing model behavior consistency, a novel formulation that preserves relations in the latent space induced by the pretrained model with relations in the latent space induced by TimeX. TimeX provides discrete attribution maps and, unlike existing interpretability methods, it learns a latent space of explanations that can be used in various ways, such as to provide landmarks to visually aggregate similar explanations and easily recognize temporal patterns. We evaluate TimeX on 8 synthetic and real-world datasets and compare its performance against state-of-the-art interpretability methods. We also conduct case studies using physiological time series. Quantitative evaluations demonstrate that TimeX achieves the highest or second-highest performance in every metric compared to baselines across all datasets. Through case studies, we show that the novel components of TimeX show potential for training faithful, interpretable models that capture the behavior of pretrained time series models.
Domain Adaptation for Time Series Under Feature and Label Shifts
Feb 06, 2023



Abstract:The transfer of models trained on labeled datasets in a source domain to unlabeled target domains is made possible by unsupervised domain adaptation (UDA). However, when dealing with complex time series models, the transferability becomes challenging due to the dynamic temporal structure that varies between domains, resulting in feature shifts and gaps in the time and frequency representations. Furthermore, tasks in the source and target domains can have vastly different label distributions, making it difficult for UDA to mitigate label shifts and recognize labels that only exist in the target domain. We present RAINCOAT, the first model for both closed-set and universal DA on complex time series. RAINCOAT addresses feature and label shifts by considering both temporal and frequency features, aligning them across domains, and correcting for misalignments to facilitate the detection of private labels. Additionally,RAINCOAT improves transferability by identifying label shifts in target domains. Our experiments with 5 datasets and 13 state-of-the-art UDA methods demonstrate that RAINCOAT can achieve an improvement in performance of up to 16.33%, and can effectively handle both closed-set and universal adaptation.
Deep Learning for Reference-Free Geolocation for Poplar Trees
Jan 31, 2023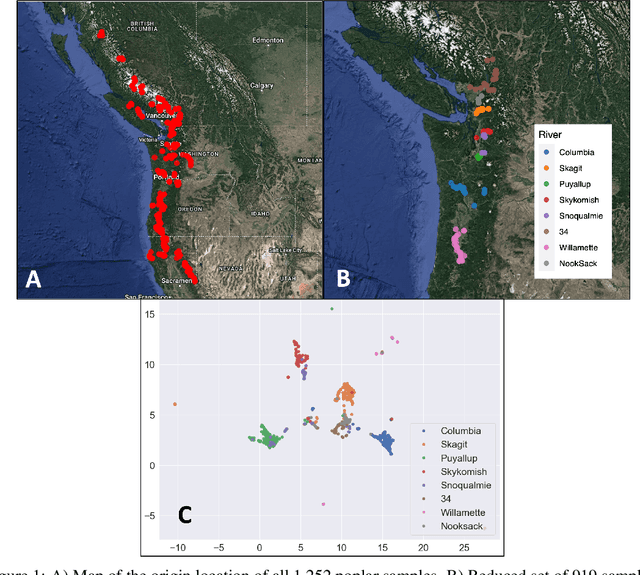
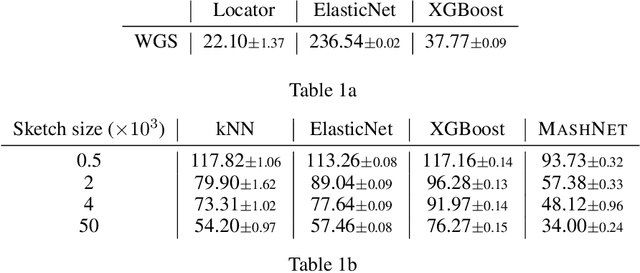
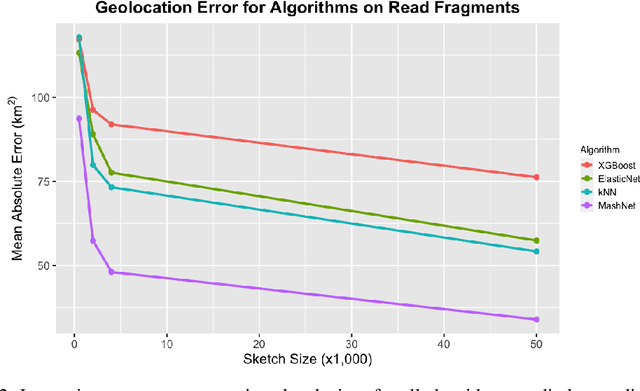
Abstract:A core task in precision agriculture is the identification of climatic and ecological conditions that are advantageous for a given crop. The most succinct approach is geolocation, which is concerned with locating the native region of a given sample based on its genetic makeup. Here, we investigate genomic geolocation of Populus trichocarpa, or poplar, which has been identified by the US Department of Energy as a fast-rotation biofuel crop to be harvested nationwide. In particular, we approach geolocation from a reference-free perspective, circumventing the need for compute-intensive processes such as variant calling and alignment. Our model, MashNet, predicts latitude and longitude for poplar trees from randomly-sampled, unaligned sequence fragments. We show that our model performs comparably to Locator, a state-of-the-art method based on aligned whole-genome sequence data. MashNet achieves an error of 34.0 km^2 compared to Locator's 22.1 km^2. MashNet allows growers to quickly and efficiently identify natural varieties that will be most productive in their growth environment based on genotype. This paper explores geolocation for precision agriculture while providing a framework and data source for further development by the machine learning community.
Evaluating Explainability for Graph Neural Networks
Aug 19, 2022



Abstract:As post hoc explanations are increasingly used to understand the behavior of graph neural networks (GNNs), it becomes crucial to evaluate the quality and reliability of GNN explanations. However, assessing the quality of GNN explanations is challenging as existing graph datasets have no or unreliable ground-truth explanations for a given task. Here, we introduce a synthetic graph data generator, ShapeGGen, which can generate a variety of benchmark datasets (e.g., varying graph sizes, degree distributions, homophilic vs. heterophilic graphs) accompanied by ground-truth explanations. Further, the flexibility to generate diverse synthetic datasets and corresponding ground-truth explanations allows us to mimic the data generated by various real-world applications. We include ShapeGGen and several real-world graph datasets into an open-source graph explainability library, GraphXAI. In addition to synthetic and real-world graph datasets with ground-truth explanations, GraphXAI provides data loaders, data processing functions, visualizers, GNN model implementations, and evaluation metrics to benchmark the performance of GNN explainability methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge