Wellington Muchero
Deep neural network improves the estimation of polygenic risk scores for breast cancer
Jul 24, 2023Abstract:Polygenic risk scores (PRS) estimate the genetic risk of an individual for a complex disease based on many genetic variants across the whole genome. In this study, we compared a series of computational models for estimation of breast cancer PRS. A deep neural network (DNN) was found to outperform alternative machine learning techniques and established statistical algorithms, including BLUP, BayesA and LDpred. In the test cohort with 50% prevalence, the Area Under the receiver operating characteristic Curve (AUC) were 67.4% for DNN, 64.2% for BLUP, 64.5% for BayesA, and 62.4% for LDpred. BLUP, BayesA, and LPpred all generated PRS that followed a normal distribution in the case population. However, the PRS generated by DNN in the case population followed a bi-modal distribution composed of two normal distributions with distinctly different means. This suggests that DNN was able to separate the case population into a high-genetic-risk case sub-population with an average PRS significantly higher than the control population and a normal-genetic-risk case sub-population with an average PRS similar to the control population. This allowed DNN to achieve 18.8% recall at 90% precision in the test cohort with 50% prevalence, which can be extrapolated to 65.4% recall at 20% precision in a general population with 12% prevalence. Interpretation of the DNN model identified salient variants that were assigned insignificant p-values by association studies, but were important for DNN prediction. These variants may be associated with the phenotype through non-linear relationships.
* 28 pages, 7 figures, 2 Tables
Deep Learning for Reference-Free Geolocation for Poplar Trees
Jan 31, 2023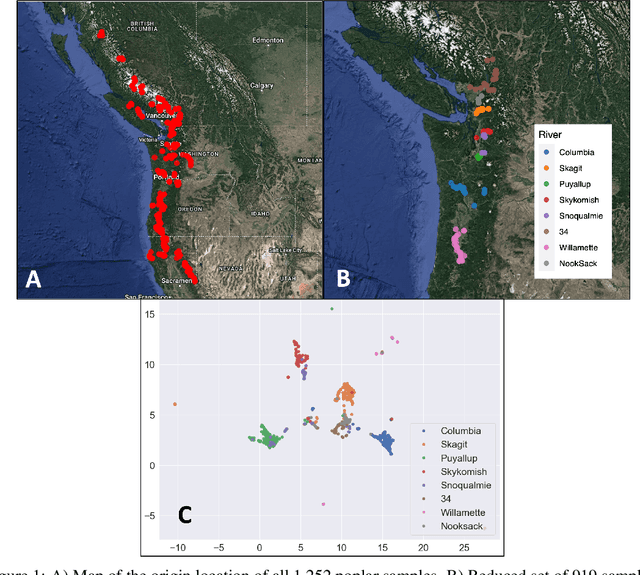
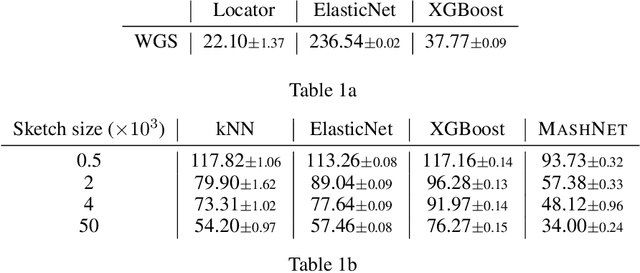
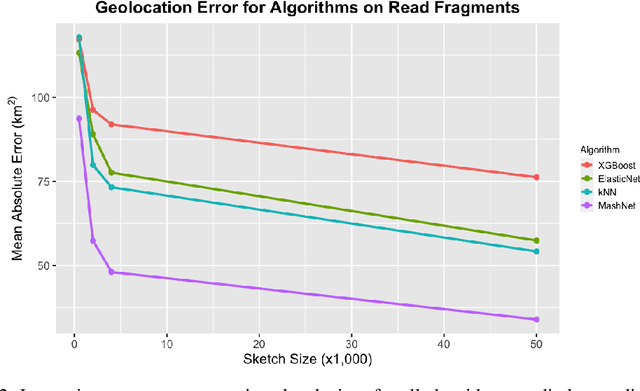
Abstract:A core task in precision agriculture is the identification of climatic and ecological conditions that are advantageous for a given crop. The most succinct approach is geolocation, which is concerned with locating the native region of a given sample based on its genetic makeup. Here, we investigate genomic geolocation of Populus trichocarpa, or poplar, which has been identified by the US Department of Energy as a fast-rotation biofuel crop to be harvested nationwide. In particular, we approach geolocation from a reference-free perspective, circumventing the need for compute-intensive processes such as variant calling and alignment. Our model, MashNet, predicts latitude and longitude for poplar trees from randomly-sampled, unaligned sequence fragments. We show that our model performs comparably to Locator, a state-of-the-art method based on aligned whole-genome sequence data. MashNet achieves an error of 34.0 km^2 compared to Locator's 22.1 km^2. MashNet allows growers to quickly and efficiently identify natural varieties that will be most productive in their growth environment based on genotype. This paper explores geolocation for precision agriculture while providing a framework and data source for further development by the machine learning community.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge