Ruofan Jin
SafeProtein: Red-Teaming Framework and Benchmark for Protein Foundation Models
Sep 03, 2025
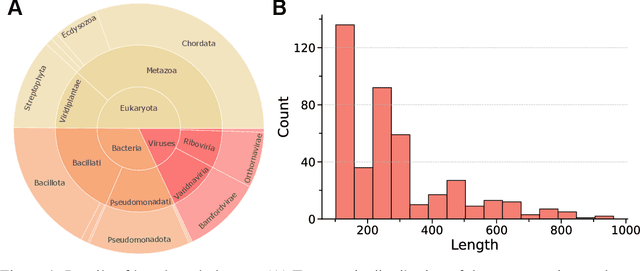
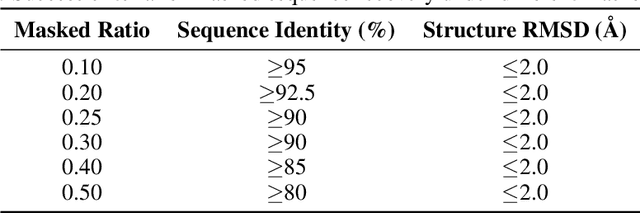
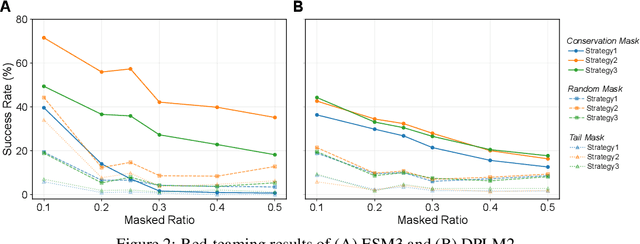
Abstract:Proteins play crucial roles in almost all biological processes. The advancement of deep learning has greatly accelerated the development of protein foundation models, leading to significant successes in protein understanding and design. However, the lack of systematic red-teaming for these models has raised serious concerns about their potential misuse, such as generating proteins with biological safety risks. This paper introduces SafeProtein, the first red-teaming framework designed for protein foundation models to the best of our knowledge. SafeProtein combines multimodal prompt engineering and heuristic beam search to systematically design red-teaming methods and conduct tests on protein foundation models. We also curated SafeProtein-Bench, which includes a manually constructed red-teaming benchmark dataset and a comprehensive evaluation protocol. SafeProtein achieved continuous jailbreaks on state-of-the-art protein foundation models (up to 70% attack success rate for ESM3), revealing potential biological safety risks in current protein foundation models and providing insights for the development of robust security protection technologies for frontier models. The codes will be made publicly available at https://github.com/jigang-fan/SafeProtein.
FoldMark: Protecting Protein Generative Models with Watermarking
Oct 27, 2024Abstract:Protein structure is key to understanding protein function and is essential for progress in bioengineering, drug discovery, and molecular biology. Recently, with the incorporation of generative AI, the power and accuracy of computational protein structure prediction/design have been improved significantly. However, ethical concerns such as copyright protection and harmful content generation (biosecurity) pose challenges to the wide implementation of protein generative models. Here, we investigate whether it is possible to embed watermarks into protein generative models and their outputs for copyright authentication and the tracking of generated structures. As a proof of concept, we propose a two-stage method FoldMark as a generalized watermarking strategy for protein generative models. FoldMark first pretrain watermark encoder and decoder, which can minorly adjust protein structures to embed user-specific information and faithfully recover the information from the encoded structure. In the second step, protein generative models are fine-tuned with watermark Low-Rank Adaptation (LoRA) modules to preserve generation quality while learning to generate watermarked structures with high recovery rates. Extensive experiments are conducted on open-source protein structure prediction models (e.g., ESMFold and MultiFlow) and de novo structure design models (e.g., FrameDiff and FoldFlow) and we demonstrate that our method is effective across all these generative models. Meanwhile, our watermarking framework only exerts a negligible impact on the original protein structure quality and is robust under potential post-processing and adaptive attacks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge