Brendan Yap
Rethinking Relational Encoding in Language Model: Pre-Training for General Sequences
Mar 18, 2021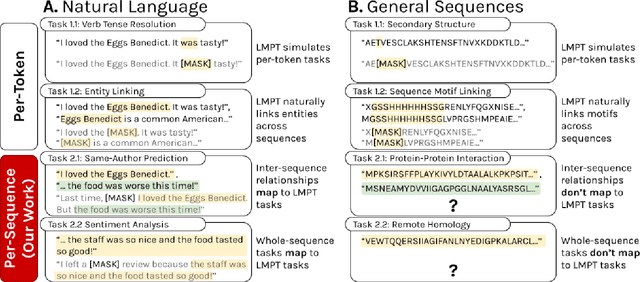
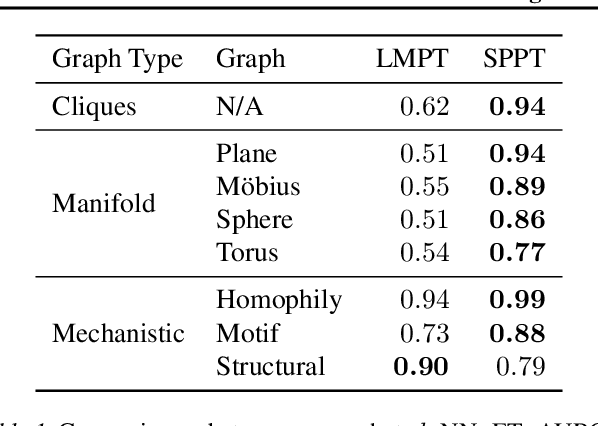
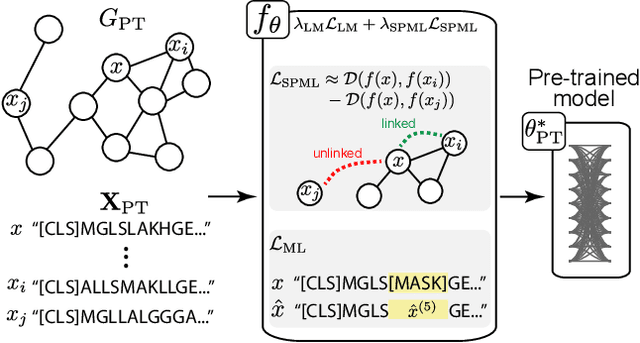
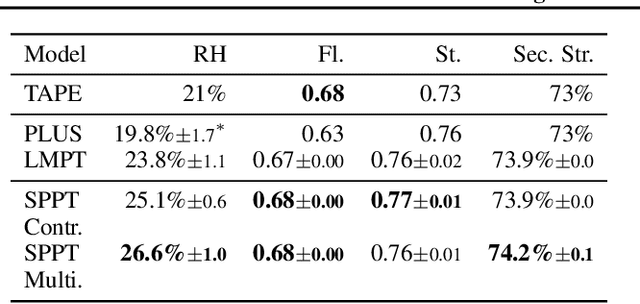
Abstract:Language model pre-training (LMPT) has achieved remarkable results in natural language understanding. However, LMPT is much less successful in non-natural language domains like protein sequences, revealing a crucial discrepancy between the various sequential domains. Here, we posit that while LMPT can effectively model per-token relations, it fails at modeling per-sequence relations in non-natural language domains. To this end, we develop a framework that couples LMPT with deep structure-preserving metric learning to produce richer embeddings than can be obtained from LMPT alone. We examine new and existing pre-training models in this framework and theoretically analyze the framework overall. We also design experiments on a variety of synthetic datasets and new graph-augmented datasets of proteins and scientific abstracts. Our approach offers notable performance improvements on downstream tasks, including prediction of protein remote homology and classification of citation intent.
Adversarial Contrastive Pre-training for Protein Sequences
Jan 31, 2021


Abstract:Recent developments in Natural Language Processing (NLP) demonstrate that large-scale, self-supervised pre-training can be extremely beneficial for downstream tasks. These ideas have been adapted to other domains, including the analysis of the amino acid sequences of proteins. However, to date most attempts on protein sequences rely on direct masked language model style pre-training. In this work, we design a new, adversarial pre-training method for proteins, extending and specializing similar advances in NLP. We show compelling results in comparison to traditional MLM pre-training, though further development is needed to ensure the gains are worth the significant computational cost.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge