Vikram Goddla
The use of large language models to enhance cancer clinical trial educational materials
Dec 02, 2024Abstract:Cancer clinical trials often face challenges in recruitment and engagement due to a lack of participant-facing informational and educational resources. This study investigated the potential of Large Language Models (LLMs), specifically GPT4, in generating patient-friendly educational content from clinical trial informed consent forms. Using data from ClinicalTrials.gov, we employed zero-shot learning for creating trial summaries and one-shot learning for developing multiple-choice questions, evaluating their effectiveness through patient surveys and crowdsourced annotation. Results showed that GPT4-generated summaries were both readable and comprehensive, and may improve patients' understanding and interest in clinical trials. The multiple-choice questions demonstrated high accuracy and agreement with crowdsourced annotators. For both resource types, hallucinations were identified that require ongoing human oversight. The findings demonstrate the potential of LLMs "out-of-the-box" to support the generation of clinical trial education materials with minimal trial-specific engineering, but implementation with a human-in-the-loop is still needed to avoid misinformation risks.
Optimal Policy Sparsification and Low Rank Decomposition for Deep Reinforcement Learning
Mar 10, 2024Abstract:Deep reinforcement learning(DRL) has shown significant promise in a wide range of applications including computer games and robotics. Yet, training DRL policies consume extraordinary computing resources resulting in dense policies which are prone to overfitting. Moreover, inference with dense DRL policies limit their practical applications, especially in edge computing. Techniques such as pruning and singular value decomposition have been used with deep learning models to achieve sparsification and model compression to limit overfitting and reduce memory consumption. However, these techniques resulted in sub-optimal performance with notable decay in rewards. $L_1$ and $L_2$ regularization techniques have been proposed for neural network sparsification and sparse auto-encoder development, but their implementation in DRL environments has not been apparent. We propose a novel $L_0$-norm-regularization technique using an optimal sparsity map to sparsify DRL policies and promote their decomposition to a lower rank without decay in rewards. We evaluated our $L_0$-norm-regularization technique across five different environments (Cartpole-v1, Acrobat-v1, LunarLander-v2, SuperMarioBros-7.1.v0 and Surgical Robot Learning) using several on-policy and off-policy algorithms. We demonstrated that the $L_0$-norm-regularized DRL policy in the SuperMarioBros environment achieved 93% sparsity and gained 70% compression when subjected to low-rank decomposition, while significantly outperforming the dense policy. Additionally, the $L_0$-norm-regularized DRL policy in the Surgical Robot Learning environment achieved a 36% sparsification and gained 46% compression when decomposed to a lower rank, while being performant. The results suggest that our custom $L_0$-norm-regularization technique for sparsification of DRL policies is a promising avenue to reduce computational resources and limit overfitting.
FocalUNETR: A Focal Transformer for Boundary-aware Segmentation of CT Images
Oct 06, 2022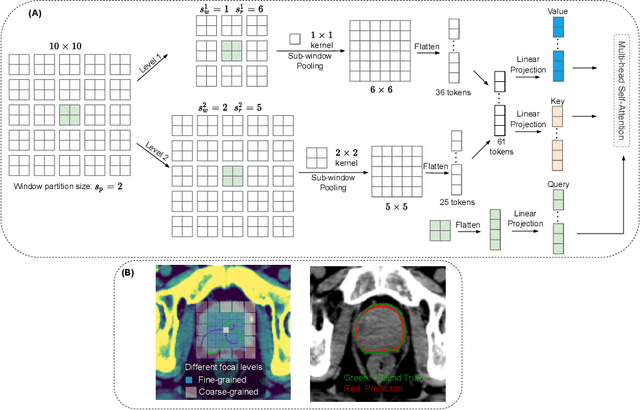
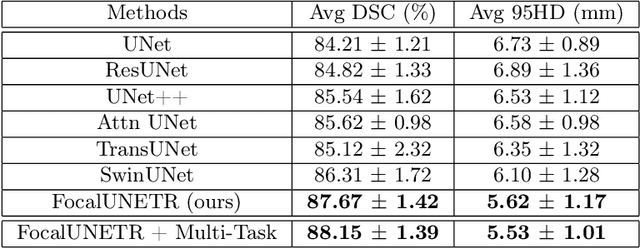
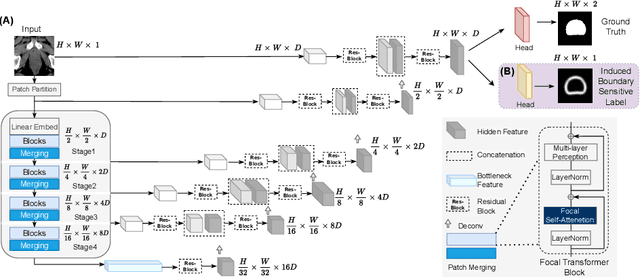

Abstract:Computed Tomography (CT) based precise prostate segmentation for treatment planning is challenging due to (1) the unclear boundary of prostate derived from CTs poor soft tissue contrast, and (2) the limitation of convolutional neural network based models in capturing long-range global context. Here we propose a focal transformer based image segmentation architecture to effectively and efficiently extract local visual features and global context from CT images. Furthermore, we design a main segmentation task and an auxiliary boundary-induced label regression task as regularization to simultaneously optimize segmentation results and mitigate the unclear boundary effect, particularly in unseen data set. Extensive experiments on a large data set of 400 prostate CT scans demonstrate the superior performance of our focal transformer to the competing methods on the prostate segmentation task.
Artificial Intelligence Solution for Effective Treatment Planning for Glioblastoma Patients
Mar 09, 2022Abstract:Glioblastomas are the most common malignant brain tumors in adults. Approximately 200000 people die each year from Glioblastoma in the world. Glioblastoma patients have a median survival of 12 months with optimal therapy and about 4 months without treatment. Glioblastomas appear as heterogeneous necrotic masses with irregular peripheral enhancement, surrounded by vasogenic edema. The current standard of care includes surgical resection, radiotherapy and chemotherapy, which require accurate segmentation of brain tumor subregions. For effective treatment planning, it is vital to identify the methylation status of the promoter of Methylguanine Methyltransferase (MGMT), a positive prognostic factor for chemotherapy. However, current methods for brain tumor segmentation are tedious, subjective and not scalable, and current techniques to determine the methylation status of MGMT promoter involve surgically invasive procedures, which are expensive and time consuming. Hence there is a pressing need to develop automated tools to segment brain tumors and non-invasive methods to predict methylation status of MGMT promoter, to facilitate better treatment planning and improve survival rate. I created an integrated diagnostics solution powered by Artificial Intelligence to automatically segment brain tumor subregions and predict MGMT promoter methylation status, using brain MRI scans. My AI solution is proven on large datasets with performance exceeding current standards and field tested with data from teaching files of local neuroradiologists. With my solution, physicians can submit brain MRI images, and get segmentation and methylation predictions in minutes, and guide brain tumor patients with effective treatment planning and ultimately improve survival time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge