David Pilcher
An Algorithmic Approach for Causal Health Equity: A Look at Race Differentials in Intensive Care Unit (ICU) Outcomes
Jan 09, 2025
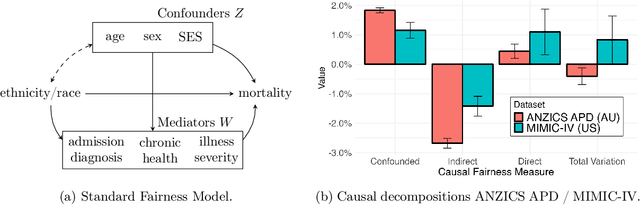
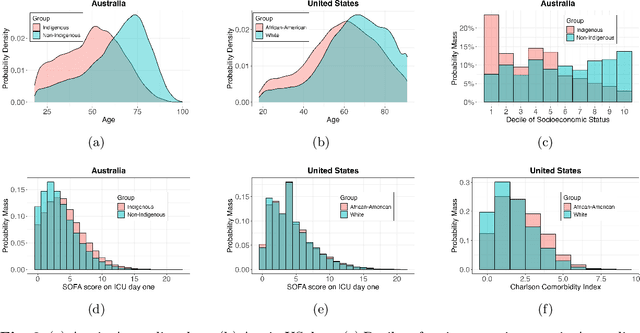

Abstract:The new era of large-scale data collection and analysis presents an opportunity for diagnosing and understanding the causes of health inequities. In this study, we describe a framework for systematically analyzing health disparities using causal inference. The framework is illustrated by investigating racial and ethnic disparities in intensive care unit (ICU) outcome between majority and minority groups in Australia (Indigenous vs. Non-Indigenous) and the United States (African-American vs. White). We demonstrate that commonly used statistical measures for quantifying inequity are insufficient, and focus on attributing the observed disparity to the causal mechanisms that generate it. We find that minority patients are younger at admission, have worse chronic health, are more likely to be admitted for urgent and non-elective reasons, and have higher illness severity. At the same time, however, we find a protective direct effect of belonging to a minority group, with minority patients showing improved survival compared to their majority counterparts, with all other variables kept equal. We demonstrate that this protective effect is related to the increased probability of being admitted to ICU, with minority patients having an increased risk of ICU admission. We also find that minority patients, while showing improved survival, are more likely to be readmitted to ICU. Thus, due to worse access to primary health care, minority patients are more likely to end up in ICU for preventable conditions, causing a reduction in the mortality rates and creating an effect that appears to be protective. Since the baseline risk of ICU admission may serve as proxy for lack of access to primary care, we developed the Indigenous Intensive Care Equity (IICE) Radar, a monitoring system for tracking the over-utilization of ICU resources by the Indigenous population of Australia across geographical areas.
Explainable Machine Learning for ICU Readmission Prediction
Sep 27, 2023



Abstract:The intensive care unit (ICU) comprises a complex hospital environment, where decisions made by clinicians have a high level of risk for the patients' lives. A comprehensive care pathway must then be followed to reduce p complications. Uncertain, competing and unplanned aspects within this environment increase the difficulty in uniformly implementing the care pathway. Readmission contributes to this pathway's difficulty, occurring when patients are admitted again to the ICU in a short timeframe, resulting in high mortality rates and high resource utilisation. Several works have tried to predict readmission through patients' medical information. Although they have some level of success while predicting readmission, those works do not properly assess, characterise and understand readmission prediction. This work proposes a standardised and explainable machine learning pipeline to model patient readmission on a multicentric database (i.e., the eICU cohort with 166,355 patients, 200,859 admissions and 6,021 readmissions) while validating it on monocentric (i.e., the MIMIC IV cohort with 382,278 patients, 523,740 admissions and 5,984 readmissions) and multicentric settings. Our machine learning pipeline achieved predictive performance in terms of the area of the receiver operating characteristic curve (AUC) up to 0.7 with a Random Forest classification model, yielding an overall good calibration and consistency on validation sets. From explanations provided by the constructed models, we could also derive a set of insightful conclusions, primarily on variables related to vital signs and blood tests (e.g., albumin, blood urea nitrogen and hemoglobin levels), demographics (e.g., age, and admission height and weight), and ICU-associated variables (e.g., unit type). These insights provide an invaluable source of information during clinicians' decision-making while discharging ICU patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge