MinJae Woo
Subgroup Performance of a Commercial Digital Breast Tomosynthesis Model for Breast Cancer Detection
Mar 17, 2025Abstract:While research has established the potential of AI models for mammography to improve breast cancer screening outcomes, there have not been any detailed subgroup evaluations performed to assess the strengths and weaknesses of commercial models for digital breast tomosynthesis (DBT) imaging. This study presents a granular evaluation of the Lunit INSIGHT DBT model on a large retrospective cohort of 163,449 screening mammography exams from the Emory Breast Imaging Dataset (EMBED). Model performance was evaluated in a binary context with various negative exam types (162,081 exams) compared against screen detected cancers (1,368 exams) as the positive class. The analysis was stratified across demographic, imaging, and pathologic subgroups to identify potential disparities. The model achieved an overall AUC of 0.91 (95% CI: 0.90-0.92) with a precision of 0.08 (95% CI: 0.08-0.08), and a recall of 0.73 (95% CI: 0.71-0.76). Performance was found to be robust across demographics, but cases with non-invasive cancers (AUC: 0.85, 95% CI: 0.83-0.87), calcifications (AUC: 0.80, 95% CI: 0.78-0.82), and dense breast tissue (AUC: 0.90, 95% CI: 0.88-0.91) were associated with significantly lower performance compared to other groups. These results highlight the need for detailed evaluation of model characteristics and vigilance in considering adoption of new tools for clinical deployment.
Benchmarking bias: Expanding clinical AI model card to incorporate bias reporting of social and non-social factors
Nov 21, 2023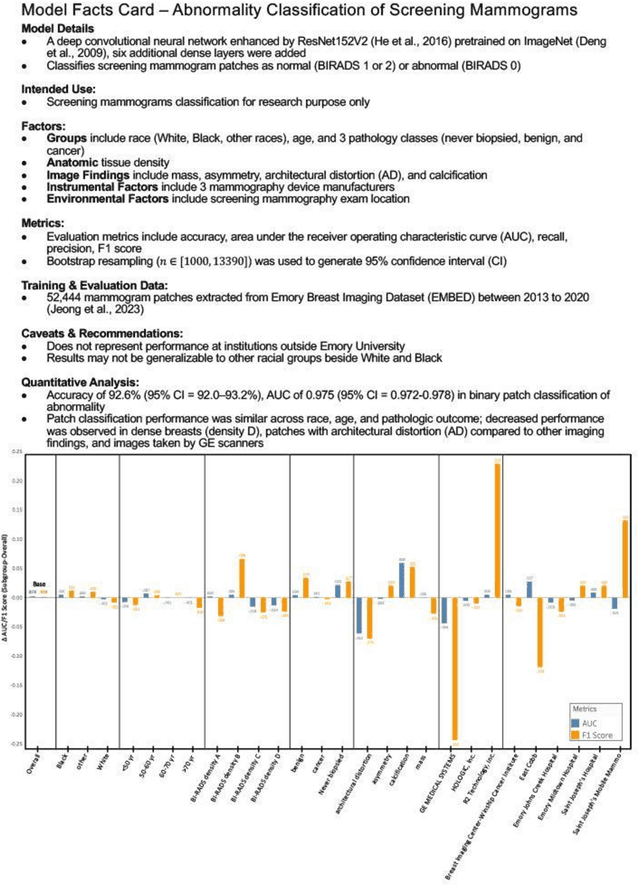
Abstract:Clinical AI model reporting cards should be expanded to incorporate a broad bias reporting of both social and non-social factors. Non-social factors consider the role of other factors, such as disease dependent, anatomic, or instrument factors on AI model bias, which are essential to ensure safe deployment.
Image Compression and Decompression Framework Based on Latent Diffusion Model for Breast Mammography
Oct 08, 2023Abstract:This research presents a novel framework for the compression and decompression of medical images utilizing the Latent Diffusion Model (LDM). The LDM represents advancement over the denoising diffusion probabilistic model (DDPM) with a potential to yield superior image quality while requiring fewer computational resources in the image decompression process. A possible application of LDM and Torchvision for image upscaling has been explored using medical image data, serving as an alternative to traditional image compression and decompression algorithms. The experimental outcomes demonstrate that this approach surpasses a conventional file compression algorithm, and convolutional neural network (CNN) models trained with decompressed files perform comparably to those trained with original image files. This approach also significantly reduces dataset size so that it can be distributed with a smaller size, and medical images take up much less space in medical devices. The research implications extend to noise reduction in lossy compression algorithms and substitute for complex wavelet-based lossless algorithms.
Performance Gaps of Artificial Intelligence Models Screening Mammography -- Towards Fair and Interpretable Models
May 08, 2023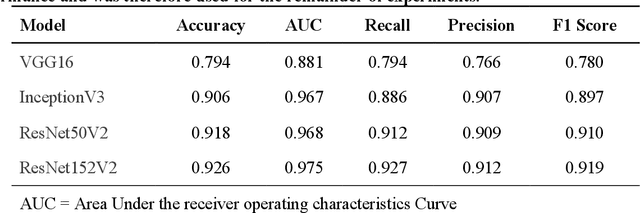
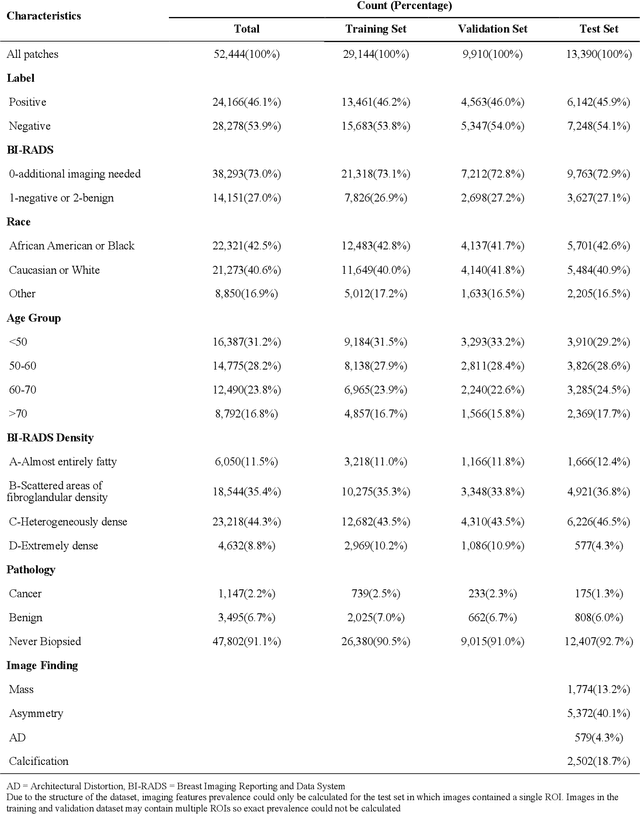
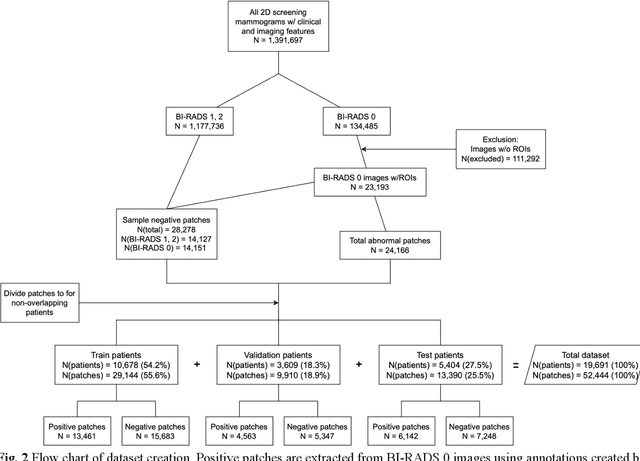
Abstract:Purpose: To analyze the demographic and imaging characteristics associated with increased risk of failure for abnormality classification in screening mammograms. Materials and Methods: This retrospective study used data from the Emory BrEast Imaging Dataset (EMBED) which includes mammograms from 115,931 patients imaged at Emory University Healthcare between 2013 to 2020. Clinical and imaging data includes Breast Imaging Reporting and Data System (BI-RADS) assessment, region of interest coordinates for abnormalities, imaging features, pathologic outcomes, and patient demographics. Multiple deep learning models were developed to distinguish between patches of abnormal tissue and randomly selected patches of normal tissue from the screening mammograms. We assessed model performance overall and within subgroups defined by age, race, pathologic outcome, and imaging characteristics to evaluate reasons for misclassifications. Results: On a test set size of 5,810 studies (13,390 patches), a ResNet152V2 model trained to classify normal versus abnormal tissue patches achieved an accuracy of 92.6% (95% CI = 92.0-93.2%), and area under the receiver operative characteristics curve 0.975 (95% CI = 0.972-0.978). Imaging characteristics associated with higher misclassifications of images include higher tissue densities (risk ratio [RR]=1.649; p=.010, BI-RADS density C and RR=2.026; p=.003, BI-RADS density D), and presence of architectural distortion (RR=1.026; p<.001). Conclusion: Even though deep learning models for abnormality classification can perform well in screening mammography, we demonstrate certain imaging features that result in worse model performance. This is the first such work to systematically evaluate breast abnormality classification by various subgroups and better-informed developers and end-users of population subgroups which are likely to experience biased model performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge