Laleh Seyyed-Kalantari
Algorithms Trained on Normal Chest X-rays Can Predict Health Insurance Types
Nov 17, 2025Abstract:Artificial intelligence is revealing what medicine never intended to encode. Deep vision models, trained on chest X-rays, can now detect not only disease but also invisible traces of social inequality. In this study, we show that state-of-the-art architectures (DenseNet121, SwinV2-B, MedMamba) can predict a patient's health insurance type, a strong proxy for socioeconomic status, from normal chest X-rays with significant accuracy (AUC around 0.67 on MIMIC-CXR-JPG, 0.68 on CheXpert). The signal persists even when age, race, and sex are controlled for, and remains detectable when the model is trained exclusively on a single racial group. Patch-based occlusion reveals that the signal is diffuse rather than localized, embedded in the upper and mid-thoracic regions. This suggests that deep networks may be internalizing subtle traces of clinical environments, equipment differences, or care pathways; learning socioeconomic segregation itself. These findings challenge the assumption that medical images are neutral biological data. By uncovering how models perceive and exploit these hidden social signatures, this work reframes fairness in medical AI: the goal is no longer only to balance datasets or adjust thresholds, but to interrogate and disentangle the social fingerprints embedded in clinical data itself.
The Impact of Unstated Norms in Bias Analysis of Language Models
Apr 07, 2024Abstract:Large language models (LLMs), trained on vast datasets, can carry biases that manifest in various forms, from overt discrimination to implicit stereotypes. One facet of bias is performance disparities in LLMs, often harming underprivileged groups, such as racial minorities. A common approach to quantifying bias is to use template-based bias probes, which explicitly state group membership (e.g. White) and evaluate if the outcome of a task, sentiment analysis for instance, is invariant to the change of group membership (e.g. change White race to Black). This approach is widely used in bias quantification. However, in this work, we find evidence of an unexpectedly overlooked consequence of using template-based probes for LLM bias quantification. We find that in doing so, text examples associated with White ethnicities appear to be classified as exhibiting negative sentiment at elevated rates. We hypothesize that the scenario arises artificially through a mismatch between the pre-training text of LLMs and the templates used to measure bias through reporting bias, unstated norms that imply group membership without explicit statement. Our finding highlights the potential misleading impact of varying group membership through explicit mention in bias quantification
Benchmarking bias: Expanding clinical AI model card to incorporate bias reporting of social and non-social factors
Nov 21, 2023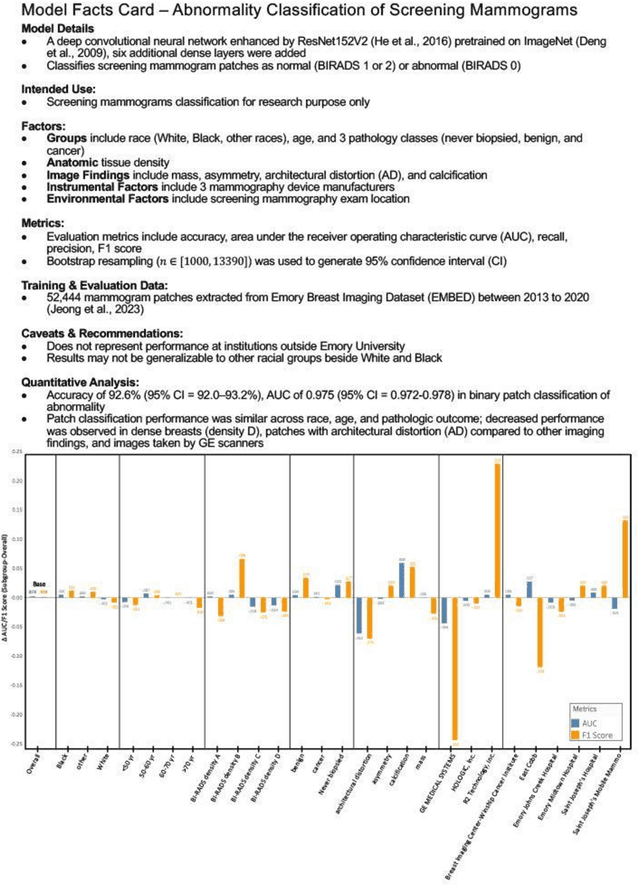
Abstract:Clinical AI model reporting cards should be expanded to incorporate a broad bias reporting of both social and non-social factors. Non-social factors consider the role of other factors, such as disease dependent, anatomic, or instrument factors on AI model bias, which are essential to ensure safe deployment.
Soft-prompt Tuning for Large Language Models to Evaluate Bias
Jun 07, 2023
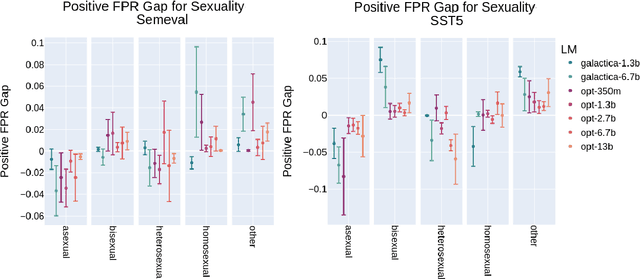
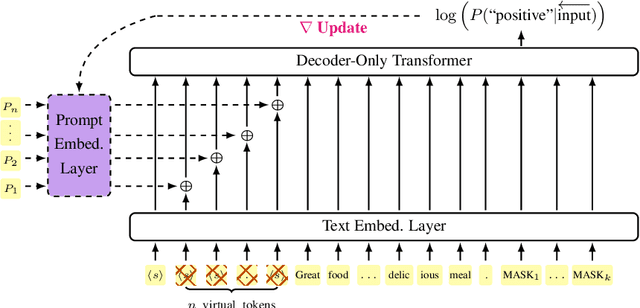
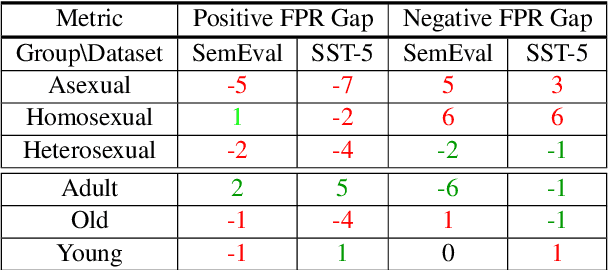
Abstract:Prompting large language models has gained immense popularity in recent years due to the advantage of producing good results even without the need for labelled data. However, this requires prompt tuning to get optimal prompts that lead to better model performances. In this paper, we explore the use of soft-prompt tuning on sentiment classification task to quantify the biases of large language models (LLMs) such as Open Pre-trained Transformers (OPT) and Galactica language model. Since these models are trained on real-world data that could be prone to bias toward certain groups of populations, it is important to identify these underlying issues. Using soft-prompts to evaluate bias gives us the extra advantage of avoiding the human-bias injection that can be caused by manually designed prompts. We check the model biases on different sensitive attributes using the group fairness (bias) and find interesting bias patterns. Since LLMs have been used in the industry in various applications, it is crucial to identify the biases before deploying these models in practice. We open-source our pipeline and encourage industry researchers to adapt our work to their use cases.
Performance Gaps of Artificial Intelligence Models Screening Mammography -- Towards Fair and Interpretable Models
May 08, 2023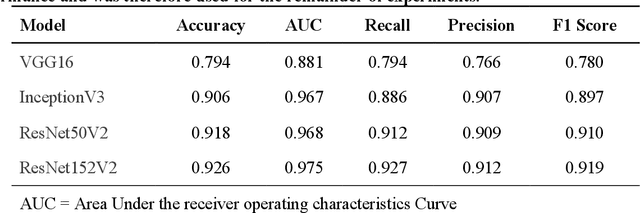
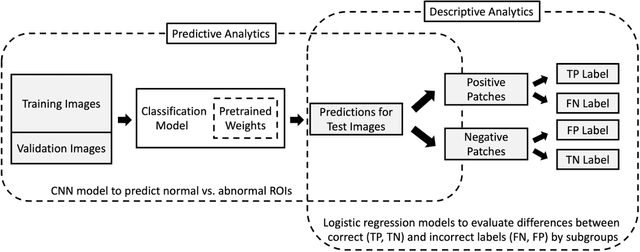
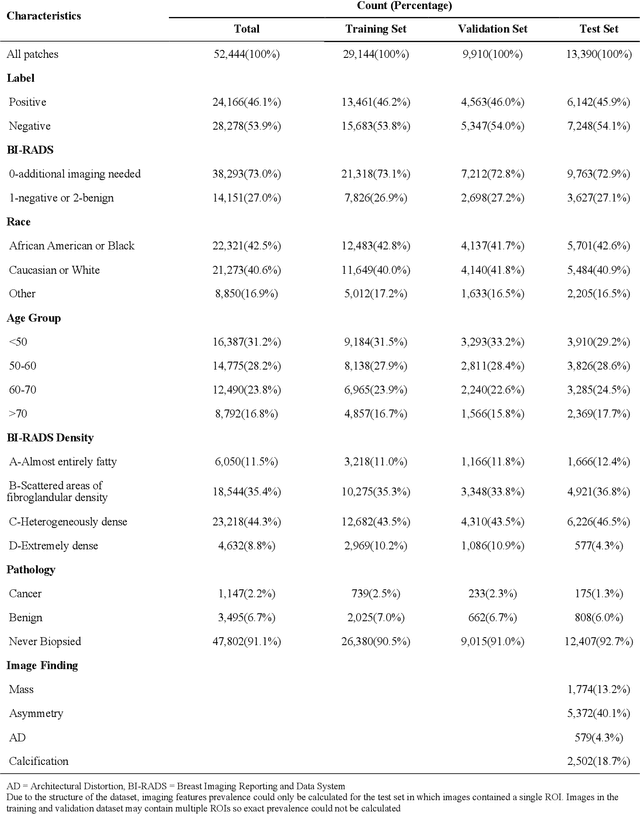
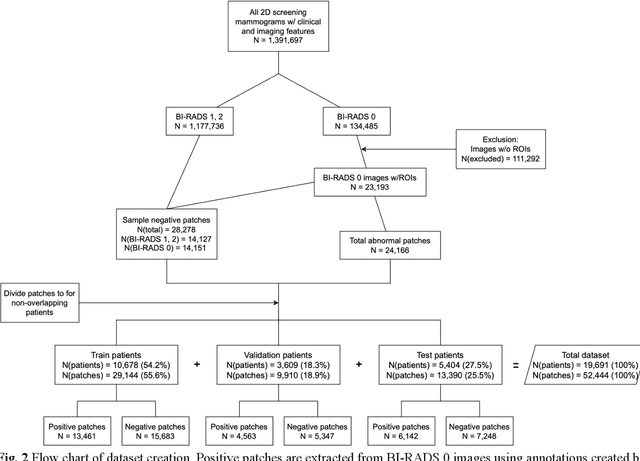
Abstract:Purpose: To analyze the demographic and imaging characteristics associated with increased risk of failure for abnormality classification in screening mammograms. Materials and Methods: This retrospective study used data from the Emory BrEast Imaging Dataset (EMBED) which includes mammograms from 115,931 patients imaged at Emory University Healthcare between 2013 to 2020. Clinical and imaging data includes Breast Imaging Reporting and Data System (BI-RADS) assessment, region of interest coordinates for abnormalities, imaging features, pathologic outcomes, and patient demographics. Multiple deep learning models were developed to distinguish between patches of abnormal tissue and randomly selected patches of normal tissue from the screening mammograms. We assessed model performance overall and within subgroups defined by age, race, pathologic outcome, and imaging characteristics to evaluate reasons for misclassifications. Results: On a test set size of 5,810 studies (13,390 patches), a ResNet152V2 model trained to classify normal versus abnormal tissue patches achieved an accuracy of 92.6% (95% CI = 92.0-93.2%), and area under the receiver operative characteristics curve 0.975 (95% CI = 0.972-0.978). Imaging characteristics associated with higher misclassifications of images include higher tissue densities (risk ratio [RR]=1.649; p=.010, BI-RADS density C and RR=2.026; p=.003, BI-RADS density D), and presence of architectural distortion (RR=1.026; p<.001). Conclusion: Even though deep learning models for abnormality classification can perform well in screening mammography, we demonstrate certain imaging features that result in worse model performance. This is the first such work to systematically evaluate breast abnormality classification by various subgroups and better-informed developers and end-users of population subgroups which are likely to experience biased model performance.
MLHOps: Machine Learning for Healthcare Operations
May 04, 2023



Abstract:Machine Learning Health Operations (MLHOps) is the combination of processes for reliable, efficient, usable, and ethical deployment and maintenance of machine learning models in healthcare settings. This paper provides both a survey of work in this area and guidelines for developers and clinicians to deploy and maintain their own models in clinical practice. We cover the foundational concepts of general machine learning operations, describe the initial setup of MLHOps pipelines (including data sources, preparation, engineering, and tools). We then describe long-term monitoring and updating (including data distribution shifts and model updating) and ethical considerations (including bias, fairness, interpretability, and privacy). This work therefore provides guidance across the full pipeline of MLHOps from conception to initial and ongoing deployment.
Reading Race: AI Recognises Patient's Racial Identity In Medical Images
Jul 21, 2021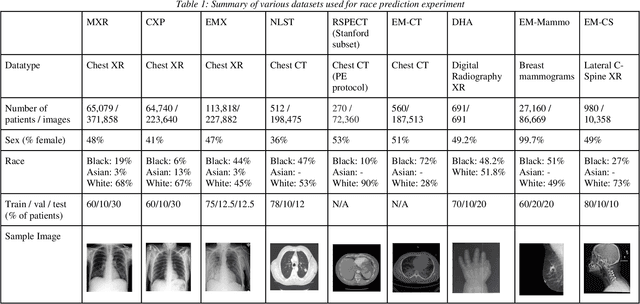
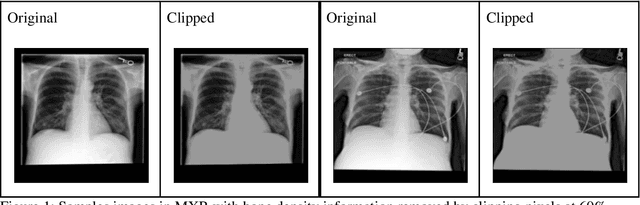
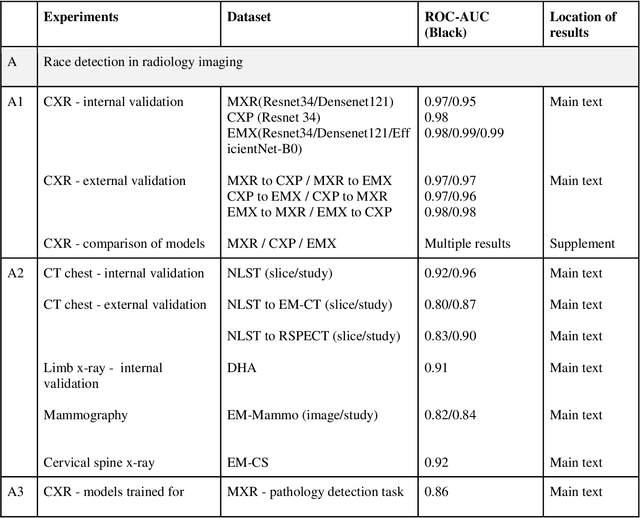
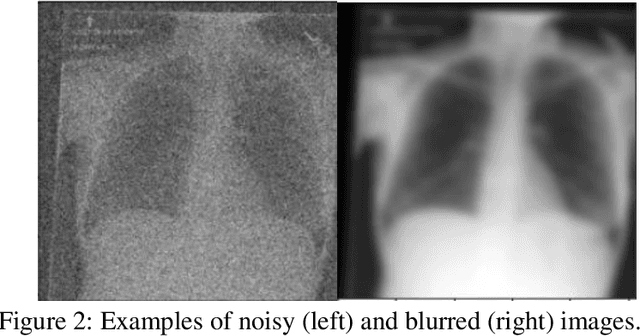
Abstract:Background: In medical imaging, prior studies have demonstrated disparate AI performance by race, yet there is no known correlation for race on medical imaging that would be obvious to the human expert interpreting the images. Methods: Using private and public datasets we evaluate: A) performance quantification of deep learning models to detect race from medical images, including the ability of these models to generalize to external environments and across multiple imaging modalities, B) assessment of possible confounding anatomic and phenotype population features, such as disease distribution and body habitus as predictors of race, and C) investigation into the underlying mechanism by which AI models can recognize race. Findings: Standard deep learning models can be trained to predict race from medical images with high performance across multiple imaging modalities. Our findings hold under external validation conditions, as well as when models are optimized to perform clinically motivated tasks. We demonstrate this detection is not due to trivial proxies or imaging-related surrogate covariates for race, such as underlying disease distribution. Finally, we show that performance persists over all anatomical regions and frequency spectrum of the images suggesting that mitigation efforts will be challenging and demand further study. Interpretation: We emphasize that model ability to predict self-reported race is itself not the issue of importance. However, our findings that AI can trivially predict self-reported race -- even from corrupted, cropped, and noised medical images -- in a setting where clinical experts cannot, creates an enormous risk for all model deployments in medical imaging: if an AI model secretly used its knowledge of self-reported race to misclassify all Black patients, radiologists would not be able to tell using the same data the model has access to.
An Empirical Framework for Domain Generalization in Clinical Settings
Apr 15, 2021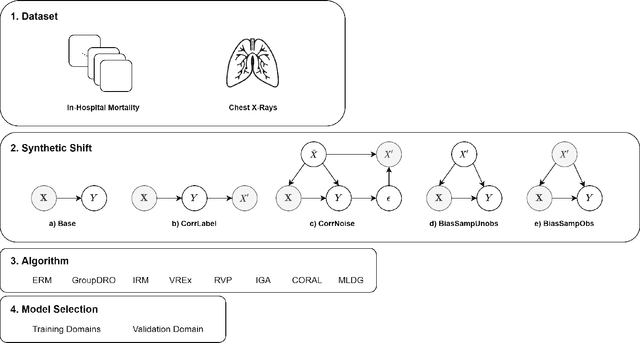
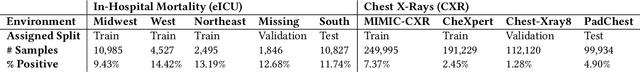
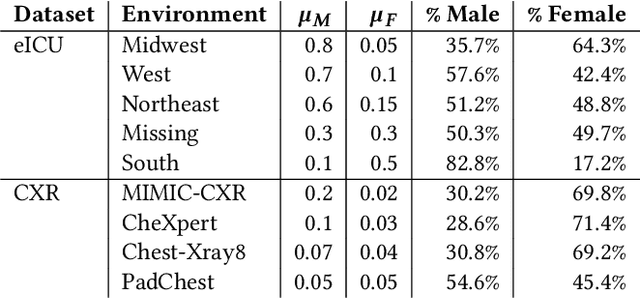
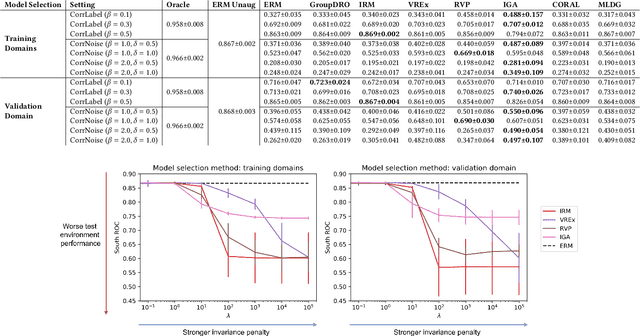
Abstract:Clinical machine learning models experience significantly degraded performance in datasets not seen during training, e.g., new hospitals or populations. Recent developments in domain generalization offer a promising solution to this problem by creating models that learn invariances across environments. In this work, we benchmark the performance of eight domain generalization methods on multi-site clinical time series and medical imaging data. We introduce a framework to induce synthetic but realistic domain shifts and sampling bias to stress-test these methods over existing non-healthcare benchmarks. We find that current domain generalization methods do not consistently achieve significant gains in out-of-distribution performance over empirical risk minimization on real-world medical imaging data, in line with prior work on general imaging datasets. However, a subset of realistic induced-shift scenarios in clinical time series data do exhibit limited performance gains. We characterize these scenarios in detail, and recommend best practices for domain generalization in the clinical setting.
Evaluating Knowledge Transfer in Neural Network for Medical Images
Sep 17, 2020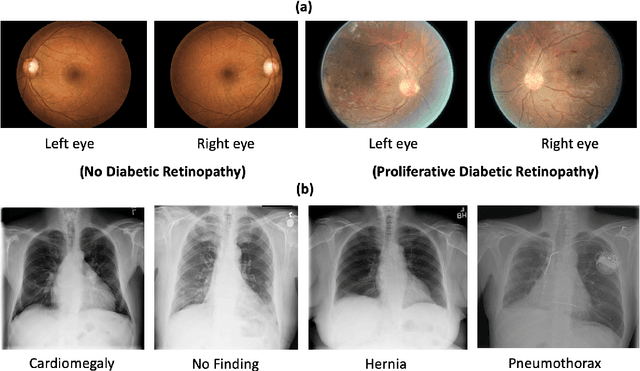
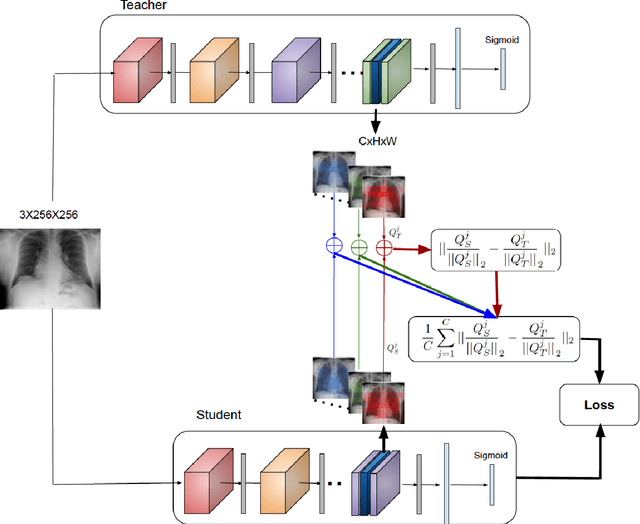
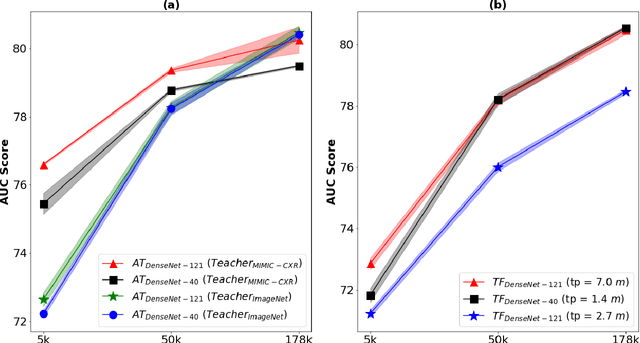
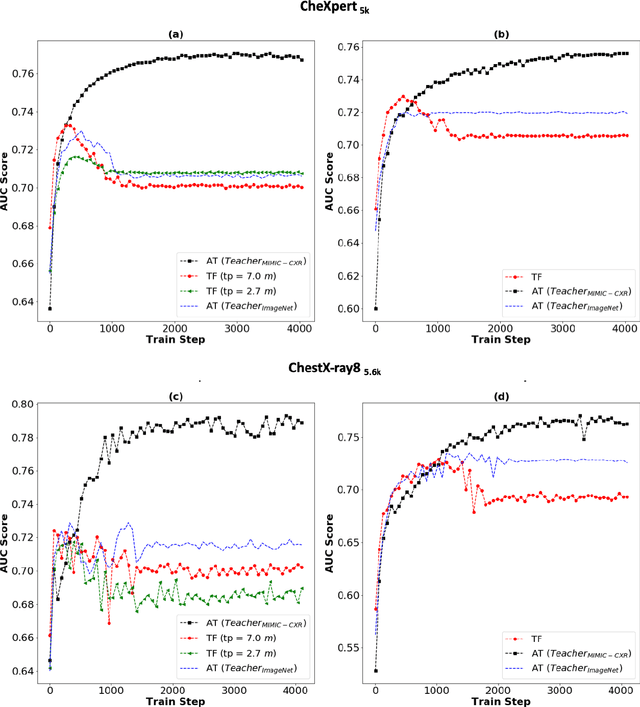
Abstract:Deep learning and knowledge transfer techniques have permeated the field of medical imaging and are considered as key approaches for revolutionizing diagnostic imaging practices. However, there are still challenges for the successful integration of deep learning into medical imaging tasks due to a lack of large annotated imaging data. To address this issue, we propose a teacher-student learning framework to transfer knowledge from a carefully pre-trained convolutional neural network (CNN) teacher to a student CNN. In this study, we explore the performance of knowledge transfer in the medical imaging setting. We investigate the proposed network's performance when the student network is trained on a small dataset (target dataset) as well as when teacher's and student's domains are distinct. The performances of the CNN models are evaluated on three medical imaging datasets including Diabetic Retinopathy, CheXpert, and ChestX-ray8. Our results indicate that the teacher-student learning framework outperforms transfer learning for small imaging datasets. Particularly, the teacher-student learning framework improves the area under the ROC Curve (AUC) of the CNN model on a small sample of CheXpert (n=5k) by 4% and on ChestX-ray8 (n=5.6k) by 9%. In addition to small training data size, we also demonstrate a clear advantage of the teacher-student learning framework in the medical imaging setting compared to transfer learning. We observe that the teacher-student network holds a great promise not only to improve the performance of diagnosis but also to reduce overfitting when the dataset is small.
CheXclusion: Fairness gaps in deep chest X-ray classifiers
Feb 14, 2020
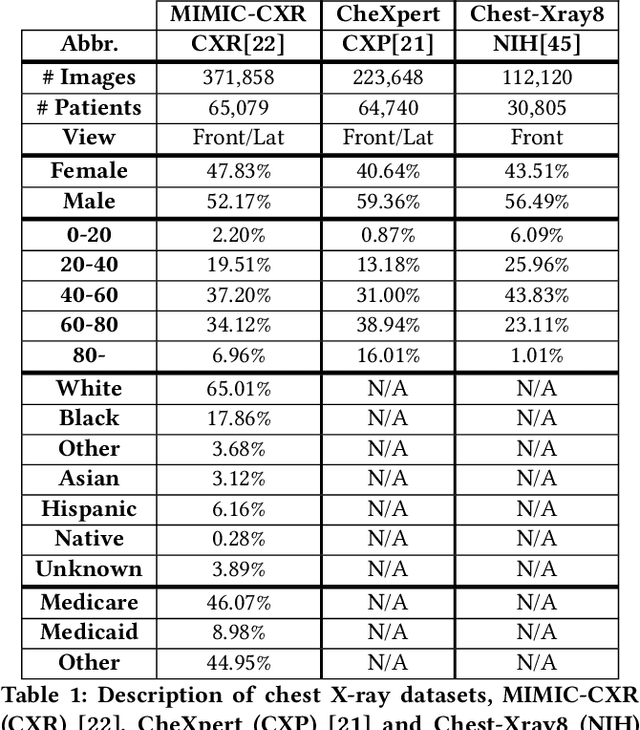
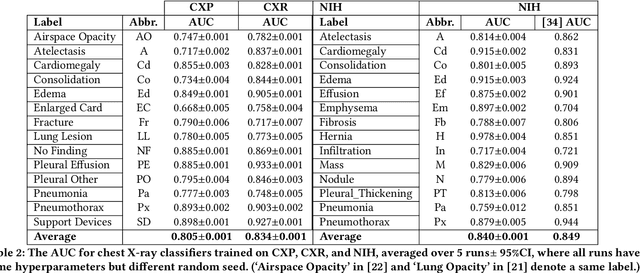
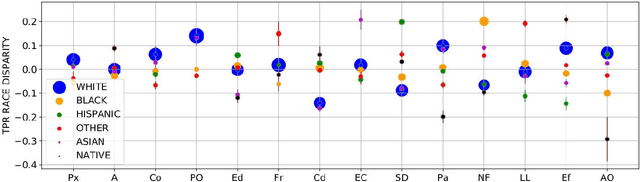
Abstract:Machine learning systems have received much attention recently for their ability to achieve expert-level performance on clinical tasks, particularly in medical imaging. Here, we examine the extent to which state-of-the-art deep learning classifiers trained to yield diagnostic labels from X-ray images are biased with respect to protected attributes. We train convolution neural networks to predict 14 diagnostic labels in three prominent public chest X-ray datasets: MIMIC-CXR, Chest-Xray8, and CheXpert. We then evaluate the TPR disparity - the difference in true positive rates (TPR) and - underdiagnosis rate - the false positive rate of a non-diagnosis - among different protected attributes such as patient sex, age, race, and insurance type. We demonstrate that TPR disparities exist in the state-of-the-art classifiers in all datasets, for all clinical tasks, and all subgroups. We find that TPR disparities are most commonly not significantly correlated with a subgroup's proportional disease burden; further, we find that some subgroups and subsection of the population are chronically underdiagnosed. Such performance disparities have real consequences as models move from papers to products, and should be carefully audited prior to deployment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge