Alistair A. Young
Potential and challenges of generative adversarial networks for super-resolution in 4D Flow MRI
Aug 20, 2025Abstract:4D Flow Magnetic Resonance Imaging (4D Flow MRI) enables non-invasive quantification of blood flow and hemodynamic parameters. However, its clinical application is limited by low spatial resolution and noise, particularly affecting near-wall velocity measurements. Machine learning-based super-resolution has shown promise in addressing these limitations, but challenges remain, not least in recovering near-wall velocities. Generative adversarial networks (GANs) offer a compelling solution, having demonstrated strong capabilities in restoring sharp boundaries in non-medical super-resolution tasks. Yet, their application in 4D Flow MRI remains unexplored, with implementation challenged by known issues such as training instability and non-convergence. In this study, we investigate GAN-based super-resolution in 4D Flow MRI. Training and validation were conducted using patient-specific cerebrovascular in-silico models, converted into synthetic images via an MR-true reconstruction pipeline. A dedicated GAN architecture was implemented and evaluated across three adversarial loss functions: Vanilla, Relativistic, and Wasserstein. Our results demonstrate that the proposed GAN improved near-wall velocity recovery compared to a non-adversarial reference (vNRMSE: 6.9% vs. 9.6%); however, that implementation specifics are critical for stable network training. While Vanilla and Relativistic GANs proved unstable compared to generator-only training (vNRMSE: 8.1% and 7.8% vs. 7.2%), a Wasserstein GAN demonstrated optimal stability and incremental improvement (vNRMSE: 6.9% vs. 7.2%). The Wasserstein GAN further outperformed the generator-only baseline at low SNR (vNRMSE: 8.7% vs. 10.7%). These findings highlight the potential of GAN-based super-resolution in enhancing 4D Flow MRI, particularly in challenging cerebrovascular regions, while emphasizing the need for careful selection of adversarial strategies.
Deep learning for temporal super-resolution 4D Flow MRI
Jan 15, 2025Abstract:4D Flow Magnetic Resonance Imaging (4D Flow MRI) is a non-invasive technique for volumetric, time-resolved blood flow quantification. However, apparent trade-offs between acquisition time, image noise, and resolution limit clinical applicability. In particular, in regions of highly transient flow, coarse temporal resolution can hinder accurate capture of physiologically relevant flow variations. To overcome these issues, post-processing techniques using deep learning have shown promising results to enhance resolution post-scan using so-called super-resolution networks. However, while super-resolution has been focusing on spatial upsampling, temporal super-resolution remains largely unexplored. The aim of this study was therefore to implement and evaluate a residual network for temporal super-resolution 4D Flow MRI. To achieve this, an existing spatial network (4DFlowNet) was re-designed for temporal upsampling, adapting input dimensions, and optimizing internal layer structures. Training and testing were performed using synthetic 4D Flow MRI data originating from patient-specific in-silico models, as well as using in-vivo datasets. Overall, excellent performance was achieved with input velocities effectively denoised and temporally upsampled, with a mean absolute error (MAE) of 1.0 cm/s in an unseen in-silico setting, outperforming deterministic alternatives (linear interpolation MAE = 2.3 cm/s, sinc interpolation MAE = 2.6 cm/s). Further, the network synthesized high-resolution temporal information from unseen low-resolution in-vivo data, with strong correlation observed at peak flow frames. As such, our results highlight the potential of utilizing data-driven neural networks for temporal super-resolution 4D Flow MRI, enabling high-frame-rate flow quantification without extending acquisition times beyond clinically acceptable limits.
Generalized super-resolution 4D Flow MRI $\unicode{x2013}$ using ensemble learning to extend across the cardiovascular system
Nov 21, 2023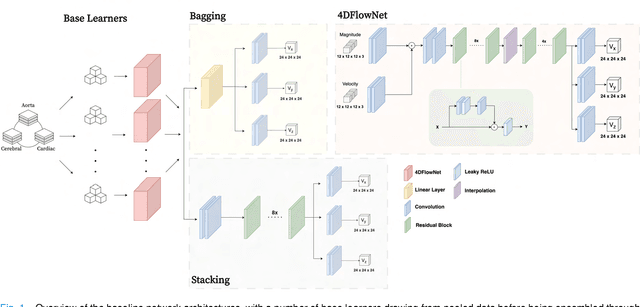
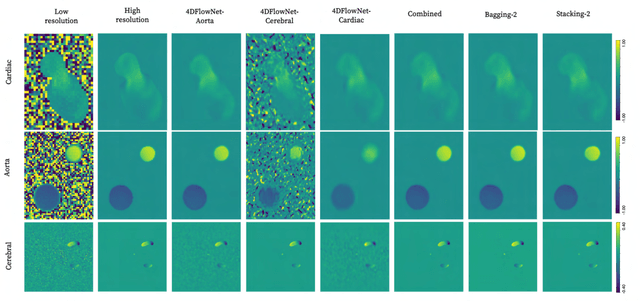
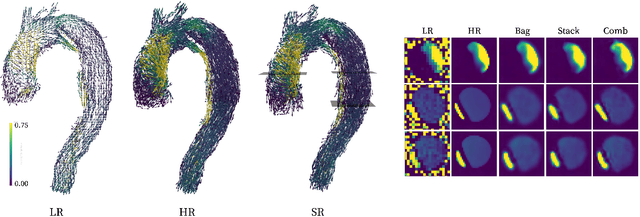
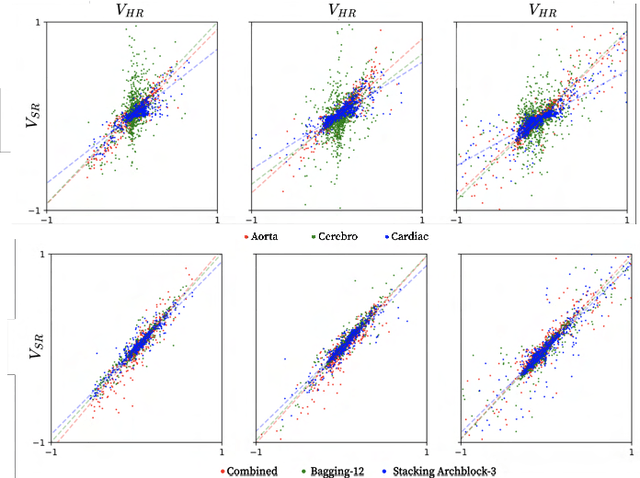
Abstract:4D Flow Magnetic Resonance Imaging (4D Flow MRI) is a non-invasive measurement technique capable of quantifying blood flow across the cardiovascular system. While practical use is limited by spatial resolution and image noise, incorporation of trained super-resolution (SR) networks has potential to enhance image quality post-scan. However, these efforts have predominantly been restricted to narrowly defined cardiovascular domains, with limited exploration of how SR performance extends across the cardiovascular system; a task aggravated by contrasting hemodynamic conditions apparent across the cardiovasculature. The aim of our study was to explore the generalizability of SR 4D Flow MRI using a combination of heterogeneous training sets and dedicated ensemble learning. With synthetic training data generated across three disparate domains (cardiac, aortic, cerebrovascular), varying convolutional base and ensemble learners were evaluated as a function of domain and architecture, quantifying performance on both in-silico and acquired in-vivo data from the same three domains. Results show that both bagging and stacking ensembling enhance SR performance across domains, accurately predicting high-resolution velocities from low-resolution input data in-silico. Likewise, optimized networks successfully recover native resolution velocities from downsampled in-vivo data, as well as show qualitative potential in generating denoised SR-images from clinical level input data. In conclusion, our work presents a viable approach for generalized SR 4D Flow MRI, with ensemble learning extending utility across various clinical areas of interest.
A Deep Learning-based Integrated Framework for Quality-aware Undersampled Cine Cardiac MRI Reconstruction and Analysis
May 02, 2022



Abstract:Cine cardiac magnetic resonance (CMR) imaging is considered the gold standard for cardiac function evaluation. However, cine CMR acquisition is inherently slow and in recent decades considerable effort has been put into accelerating scan times without compromising image quality or the accuracy of derived results. In this paper, we present a fully-automated, quality-controlled integrated framework for reconstruction, segmentation and downstream analysis of undersampled cine CMR data. The framework enables active acquisition of radial k-space data, in which acquisition can be stopped as soon as acquired data are sufficient to produce high quality reconstructions and segmentations. This results in reduced scan times and automated analysis, enabling robust and accurate estimation of functional biomarkers. To demonstrate the feasibility of the proposed approach, we perform realistic simulations of radial k-space acquisitions on a dataset of subjects from the UK Biobank and present results on in-vivo cine CMR k-space data collected from healthy subjects. The results demonstrate that our method can produce quality-controlled images in a mean scan time reduced from 12 to 4 seconds per slice, and that image quality is sufficient to allow clinically relevant parameters to be automatically estimated to within 5% mean absolute difference.
Surface Vision Transformers: Flexible Attention-Based Modelling of Biomedical Surfaces
Apr 07, 2022



Abstract:Recent state-of-the-art performances of Vision Transformers (ViT) in computer vision tasks demonstrate that a general-purpose architecture, which implements long-range self-attention, could replace the local feature learning operations of convolutional neural networks. In this paper, we extend ViTs to surfaces by reformulating the task of surface learning as a sequence-to-sequence learning problem, by proposing patching mechanisms for general surface meshes. Sequences of patches are then processed by a transformer encoder and used for classification or regression. We validate our method on a range of different biomedical surface domains and tasks: brain age prediction in the developing Human Connectome Project (dHCP), fluid intelligence prediction in the Human Connectome Project (HCP), and coronary artery calcium score classification using surfaces from the Scottish Computed Tomography of the Heart (SCOT-HEART) dataset, and investigate the impact of pretraining and data augmentation on model performance. Results suggest that Surface Vision Transformers (SiT) demonstrate consistent improvement over geometric deep learning methods for brain age and fluid intelligence prediction and achieve comparable performance on calcium score classification to standard metrics used in clinical practice. Furthermore, analysis of transformer attention maps offers clear and individualised predictions of the features driving each task. Code is available on Github: https://github.com/metrics-lab/surface-vision-transformers
Whole Heart Anatomical Refinement from CCTA using Extrapolation and Parcellation
Nov 18, 2021



Abstract:Coronary computed tomography angiography (CCTA) provides detailed an-atomical information on all chambers of the heart. Existing segmentation tools can label the gross anatomy, but addition of application-specific labels can require detailed and often manual refinement. We developed a U-Net based framework to i) extrapolate a new label from existing labels, and ii) parcellate one label into multiple labels, both using label-to-label mapping, to create a desired segmentation that could then be learnt directly from the image (image- to-label mapping). This approach only required manual correction in a small subset of cases (80 for extrapolation, 50 for parcella-tion, compared with 260 for initial labels). An initial 6-label segmentation (left ventricle, left ventricular myocardium, right ventricle, left atrium, right atrium and aorta) was refined to a 10-label segmentation that added a label for the pulmonary artery and divided the left atrium label into body, left and right veins and appendage components. The final method was tested using 30 cases, 10 each from Philips, Siemens and Toshiba scanners. In addition to the new labels, the median Dice scores were improved for all the initial 6 labels to be above 95% in the 10-label segmentation, e.g. from 91% to 97% for the left atrium body and from 92% to 96% for the right ventricle. This method provides a simple framework for flexible refinement of anatomical labels. The code and executables are available at cemrg.com.
Fully Automated Myocardial Strain Estimation from CMR Tagged Images using a Deep Learning Framework in the UK Biobank
Apr 15, 2020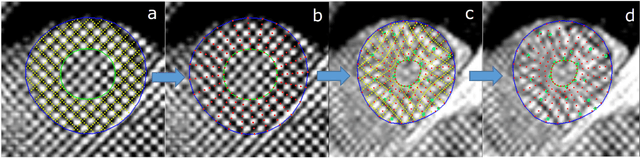
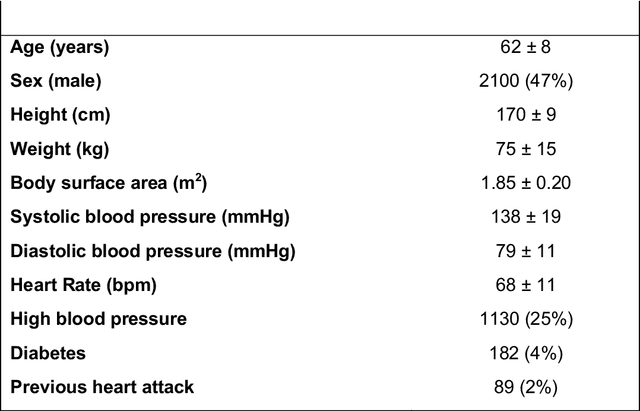
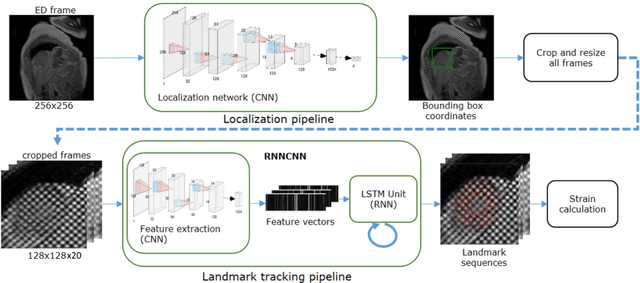
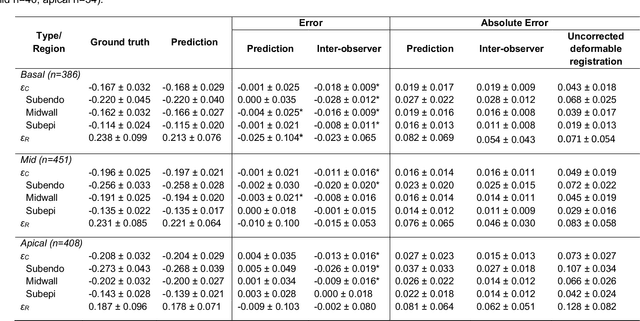
Abstract:Purpose: To demonstrate the feasibility and performance of a fully automated deep learning framework to estimate myocardial strain from short-axis cardiac magnetic resonance tagged images. Methods and Materials: In this retrospective cross-sectional study, 4508 cases from the UK Biobank were split randomly into 3244 training and 812 validation cases, and 452 test cases. Ground truth myocardial landmarks were defined and tracked by manual initialization and correction of deformable image registration using previously validated software with five readers. The fully automatic framework consisted of 1) a convolutional neural network (CNN) for localization, and 2) a combination of a recurrent neural network (RNN) and a CNN to detect and track the myocardial landmarks through the image sequence for each slice. Radial and circumferential strain were then calculated from the motion of the landmarks and averaged on a slice basis. Results: Within the test set, myocardial end-systolic circumferential Green strain errors were -0.001 +/- 0.025, -0.001 +/- 0.021, and 0.004 +/- 0.035 in basal, mid, and apical slices respectively (mean +/- std. dev. of differences between predicted and manual strain). The framework reproduced significant reductions in circumferential strain in diabetics, hypertensives, and participants with previous heart attack. Typical processing time was ~260 frames (~13 slices) per second on an NVIDIA Tesla K40 with 12GB RAM, compared with 6-8 minutes per slice for the manual analysis. Conclusions: The fully automated RNNCNN framework for analysis of myocardial strain enabled unbiased strain evaluation in a high-throughput workflow, with similar ability to distinguish impairment due to diabetes, hypertension, and previous heart attack.
* accepted in Radiology Cardiothoracic Imaging
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge