Michelle C. Williams
Exploring Multimodal Large Language Models for Radiology Report Error-checking
Dec 20, 2023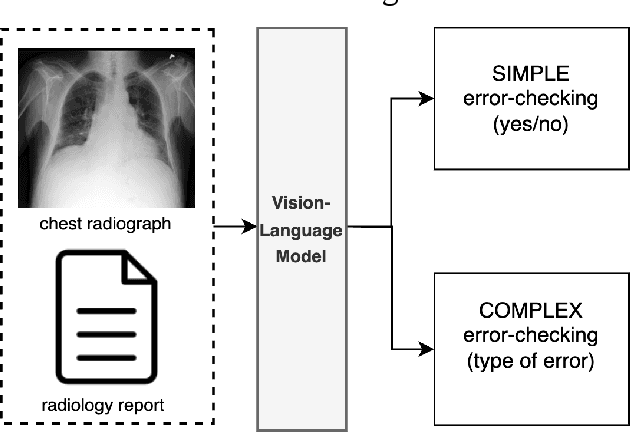
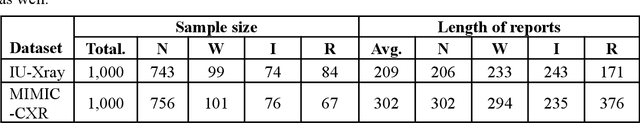

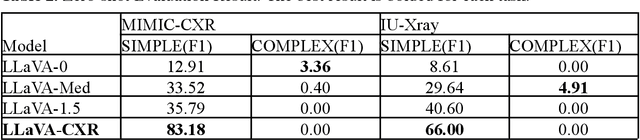
Abstract:This paper proposes one of the first clinical applications of multimodal large language models (LLMs) as an assistant for radiologists to check errors in their reports. We created an evaluation dataset from two real-world radiology datasets (MIMIC-CXR and IU-Xray), with 1,000 subsampled reports each. A subset of original reports was modified to contain synthetic errors by introducing various type of mistakes. The evaluation contained two difficulty levels: SIMPLE for binary error-checking and COMPLEX for identifying error types. LLaVA (Large Language and Visual Assistant) variant models, including our instruction-tuned model, were used for the evaluation. Additionally, a domain expert evaluation was conducted on a small test set. At the SIMPLE level, the LLaVA v1.5 model outperformed other publicly available models. Instruction tuning significantly enhanced performance by 47.4% and 25.4% on MIMIC-CXR and IU-Xray data, respectively. The model also surpassed the domain experts accuracy in the MIMIC-CXR dataset by 1.67%. Notably, among the subsets (N=21) of the test set where a clinician did not achieve the correct conclusion, the LLaVA ensemble mode correctly identified 71.4% of these cases. This study marks a promising step toward utilizing multi-modal LLMs to enhance diagnostic accuracy in radiology. The ensemble model demonstrated comparable performance to clinicians, even capturing errors overlooked by humans. Nevertheless, future work is needed to improve the model ability to identify the types of inconsistency.
Surface Vision Transformers: Flexible Attention-Based Modelling of Biomedical Surfaces
Apr 07, 2022



Abstract:Recent state-of-the-art performances of Vision Transformers (ViT) in computer vision tasks demonstrate that a general-purpose architecture, which implements long-range self-attention, could replace the local feature learning operations of convolutional neural networks. In this paper, we extend ViTs to surfaces by reformulating the task of surface learning as a sequence-to-sequence learning problem, by proposing patching mechanisms for general surface meshes. Sequences of patches are then processed by a transformer encoder and used for classification or regression. We validate our method on a range of different biomedical surface domains and tasks: brain age prediction in the developing Human Connectome Project (dHCP), fluid intelligence prediction in the Human Connectome Project (HCP), and coronary artery calcium score classification using surfaces from the Scottish Computed Tomography of the Heart (SCOT-HEART) dataset, and investigate the impact of pretraining and data augmentation on model performance. Results suggest that Surface Vision Transformers (SiT) demonstrate consistent improvement over geometric deep learning methods for brain age and fluid intelligence prediction and achieve comparable performance on calcium score classification to standard metrics used in clinical practice. Furthermore, analysis of transformer attention maps offers clear and individualised predictions of the features driving each task. Code is available on Github: https://github.com/metrics-lab/surface-vision-transformers
Whole Heart Anatomical Refinement from CCTA using Extrapolation and Parcellation
Nov 18, 2021



Abstract:Coronary computed tomography angiography (CCTA) provides detailed an-atomical information on all chambers of the heart. Existing segmentation tools can label the gross anatomy, but addition of application-specific labels can require detailed and often manual refinement. We developed a U-Net based framework to i) extrapolate a new label from existing labels, and ii) parcellate one label into multiple labels, both using label-to-label mapping, to create a desired segmentation that could then be learnt directly from the image (image- to-label mapping). This approach only required manual correction in a small subset of cases (80 for extrapolation, 50 for parcella-tion, compared with 260 for initial labels). An initial 6-label segmentation (left ventricle, left ventricular myocardium, right ventricle, left atrium, right atrium and aorta) was refined to a 10-label segmentation that added a label for the pulmonary artery and divided the left atrium label into body, left and right veins and appendage components. The final method was tested using 30 cases, 10 each from Philips, Siemens and Toshiba scanners. In addition to the new labels, the median Dice scores were improved for all the initial 6 labels to be above 95% in the 10-label segmentation, e.g. from 91% to 97% for the left atrium body and from 92% to 96% for the right ventricle. This method provides a simple framework for flexible refinement of anatomical labels. The code and executables are available at cemrg.com.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge