Paul Taylor
A computationally frugal open-source foundation model for thoracic disease detection in lung cancer screening programs
Jul 02, 2025Abstract:Low-dose computed tomography (LDCT) imaging employed in lung cancer screening (LCS) programs is increasing in uptake worldwide. LCS programs herald a generational opportunity to simultaneously detect cancer and non-cancer-related early-stage lung disease. Yet these efforts are hampered by a shortage of radiologists to interpret scans at scale. Here, we present TANGERINE, a computationally frugal, open-source vision foundation model for volumetric LDCT analysis. Designed for broad accessibility and rapid adaptation, TANGERINE can be fine-tuned off the shelf for a wide range of disease-specific tasks with limited computational resources and training data. Relative to models trained from scratch, TANGERINE demonstrates fast convergence during fine-tuning, thereby requiring significantly fewer GPU hours, and displays strong label efficiency, achieving comparable or superior performance with a fraction of fine-tuning data. Pretrained using self-supervised learning on over 98,000 thoracic LDCTs, including the UK's largest LCS initiative to date and 27 public datasets, TANGERINE achieves state-of-the-art performance across 14 disease classification tasks, including lung cancer and multiple respiratory diseases, while generalising robustly across diverse clinical centres. By extending a masked autoencoder framework to 3D imaging, TANGERINE offers a scalable solution for LDCT analysis, departing from recent closed, resource-intensive models by combining architectural simplicity, public availability, and modest computational requirements. Its accessible, open-source lightweight design lays the foundation for rapid integration into next-generation medical imaging tools that could transform LCS initiatives, allowing them to pivot from a singular focus on lung cancer detection to comprehensive respiratory disease management in high-risk populations.
Minuscule Cell Detection in AS-OCT Images with Progressive Field-of-View Focusing
Mar 15, 2025Abstract:Anterior Segment Optical Coherence Tomography (AS-OCT) is an emerging imaging technique with great potential for diagnosing anterior uveitis, a vision-threatening ocular inflammatory condition. A hallmark of this condition is the presence of inflammatory cells in the eye's anterior chamber, and detecting these cells using AS-OCT images has attracted research interest. While recent efforts aim to replace manual cell detection with automated computer vision approaches, detecting extremely small (minuscule) objects in high-resolution images, such as AS-OCT, poses substantial challenges: (1) each cell appears as a minuscule particle, representing less than 0.005\% of the image, making the detection difficult, and (2) OCT imaging introduces pixel-level noise that can be mistaken for cells, leading to false positive detections. To overcome these challenges, we propose a minuscule cell detection framework through a progressive field-of-view focusing strategy. This strategy systematically refines the detection scope from the whole image to a target region where cells are likely to be present, and further to minuscule regions potentially containing individual cells. Our framework consists of two modules. First, a Field-of-Focus module uses a vision foundation model to segment the target region. Subsequently, a Fine-grained Object Detection module introduces a specialized Minuscule Region Proposal followed by a Spatial Attention Network to distinguish individual cells from noise within the segmented region. Experimental results demonstrate that our framework outperforms state-of-the-art methods for cell detection, providing enhanced efficacy for clinical applications. Our code is publicly available at: https://github.com/joeybyc/MCD.
Medical Image Quality Assessment based on Probability of Necessity and Sufficiency
Oct 10, 2024



Abstract:Medical image quality assessment (MIQA) is essential for reliable medical image analysis. While deep learning has shown promise in this field, current models could be misled by spurious correlations learned from data and struggle with out-of-distribution (OOD) scenarios. To that end, we propose an MIQA framework based on a concept from causal inference: Probability of Necessity and Sufficiency (PNS). PNS measures how likely a set of features is to be both necessary (always present for an outcome) and sufficient (capable of guaranteeing an outcome) for a particular result. Our approach leverages this concept by learning hidden features from medical images with high PNS values for quality prediction. This encourages models to capture more essential predictive information, enhancing their robustness to OOD scenarios. We evaluate our framework on an Anterior Segment Optical Coherence Tomography (AS-OCT) dataset for the MIQA task and experimental results demonstrate the effectiveness of our framework.
Advancing Cell Detection in Anterior Segment Optical Coherence Tomography Images
Jun 25, 2024Abstract:Anterior uveitis, a common form of eye inflammation, can lead to permanent vision loss if not promptly diagnosed. Monitoring this condition involves quantifying inflammatory cells in the anterior chamber (AC) of the eye, which can be captured using Anterior Segment Optical Coherence Tomography (AS-OCT). However, manually identifying cells in AS-OCT images is time-consuming and subjective. Moreover, existing automated approaches may have limitations in both the effectiveness of detecting cells and the reliability of their detection results. To address these challenges, we propose an automated framework to detect cells in the AS-OCT images. This framework consists of a zero-shot chamber segmentation module and a cell detection module. The first module segments the AC area in the image without requiring human-annotated training data. Subsequently, the second module identifies individual cells within the segmented AC region. Through experiments, our framework demonstrates superior performance compared to current state-of-the-art methods for both AC segmentation and cell detection tasks. Notably, we find that previous cell detection approaches could suffer from low recall, potentially overlooking a significant number of cells. In contrast, our framework offers an improved solution, which could benefit the diagnosis and study of anterior uveitis. Our code for cell detection is publicly available at: https://github.com/joeybyc/cell_detection.
Privacy Preserved Blood Glucose Level Cross-Prediction: An Asynchronous Decentralized Federated Learning Approach
Jun 21, 2024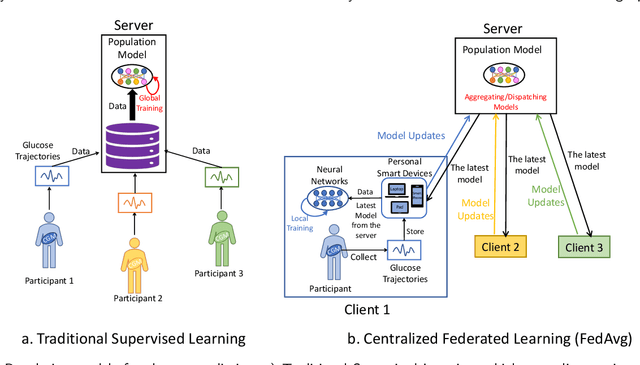
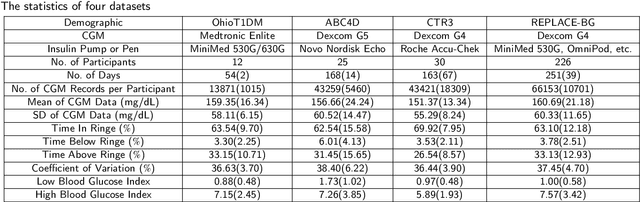
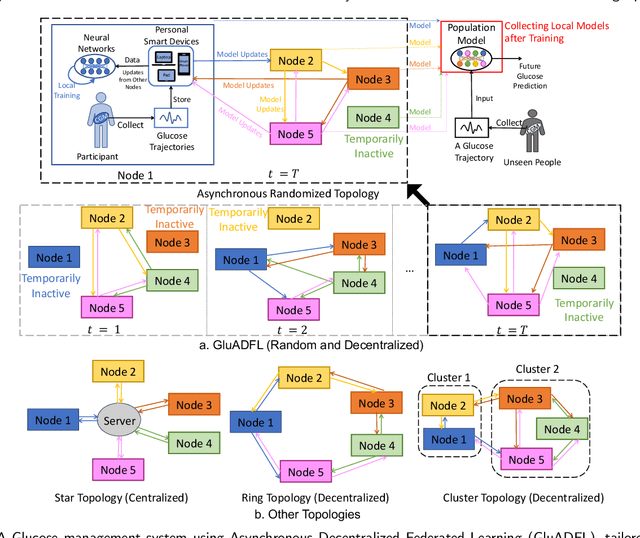
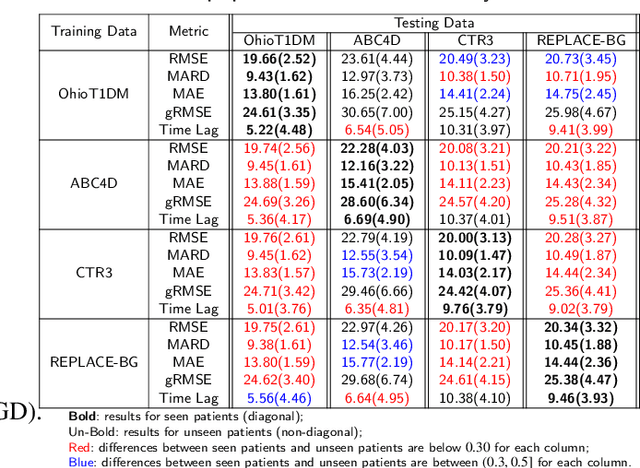
Abstract:Newly diagnosed Type 1 Diabetes (T1D) patients often struggle to obtain effective Blood Glucose (BG) prediction models due to the lack of sufficient BG data from Continuous Glucose Monitoring (CGM), presenting a significant "cold start" problem in patient care. Utilizing population models to address this challenge is a potential solution, but collecting patient data for training population models in a privacy-conscious manner is challenging, especially given that such data is often stored on personal devices. Considering the privacy protection and addressing the "cold start" problem in diabetes care, we propose "GluADFL", blood Glucose prediction by Asynchronous Decentralized Federated Learning. We compared GluADFL with eight baseline methods using four distinct T1D datasets, comprising 298 participants, which demonstrated its superior performance in accurately predicting BG levels for cross-patient analysis. Furthermore, patients' data might be stored and shared across various communication networks in GluADFL, ranging from highly interconnected (e.g., random, performs the best among others) to more structured topologies (e.g., cluster and ring), suitable for various social networks. The asynchronous training framework supports flexible participation. By adjusting the ratios of inactive participants, we found it remains stable if less than 70% are inactive. Our results confirm that GluADFL offers a practical, privacy-preserving solution for BG prediction in T1D, significantly enhancing the quality of diabetes management.
A Data-driven Approach for Rapid Detection of Aeroelastic Modes from Flutter Flight Test Based on Limited Sensor Measurements
Mar 18, 2024



Abstract:Flutter flight test involves the evaluation of the airframes aeroelastic stability by applying artificial excitation on the aircraft lifting surfaces. The subsequent responses are captured and analyzed to extract the frequencies and damping characteristics of the system. However, noise contamination, turbulence, non-optimal excitation of modes, and sensor malfunction in one or more sensors make it time-consuming and corrupt the extraction process. In order to expedite the process of identifying and analyzing aeroelastic modes, this study implements a time-delay embedded Dynamic Mode Decomposition technique. This approach is complemented by Robust Principal Component Analysis methodology, and a sparsity promoting criterion which enables the automatic and optimal selection of sparse modes. The anonymized flutter flight test data, provided by the fifth author of this research paper, is utilized in this implementation. The methodology assumes no knowledge of the input excitation, only deals with the responses captured by accelerometer channels, and rapidly identifies the aeroelastic modes. By incorporating a compressed sensing algorithm, the methodology gains the ability to identify aeroelastic modes, even when the number of available sensors is limited. This augmentation greatly enhances the methodology's robustness and effectiveness, making it an excellent choice for real-time implementation during flutter test campaigns.
GARNN: An Interpretable Graph Attentive Recurrent Neural Network for Predicting Blood Glucose Levels via Multivariate Time Series
Feb 26, 2024



Abstract:Accurate prediction of future blood glucose (BG) levels can effectively improve BG management for people living with diabetes, thereby reducing complications and improving quality of life. The state of the art of BG prediction has been achieved by leveraging advanced deep learning methods to model multi-modal data, i.e., sensor data and self-reported event data, organised as multi-variate time series (MTS). However, these methods are mostly regarded as ``black boxes'' and not entirely trusted by clinicians and patients. In this paper, we propose interpretable graph attentive recurrent neural networks (GARNNs) to model MTS, explaining variable contributions via summarizing variable importance and generating feature maps by graph attention mechanisms instead of post-hoc analysis. We evaluate GARNNs on four datasets, representing diverse clinical scenarios. Upon comparison with twelve well-established baseline methods, GARNNs not only achieve the best prediction accuracy but also provide high-quality temporal interpretability, in particular for postprandial glucose levels as a result of corresponding meal intake and insulin injection. These findings underline the potential of GARNN as a robust tool for improving diabetes care, bridging the gap between deep learning technology and real-world healthcare solutions.
Exploring Multimodal Large Language Models for Radiology Report Error-checking
Dec 20, 2023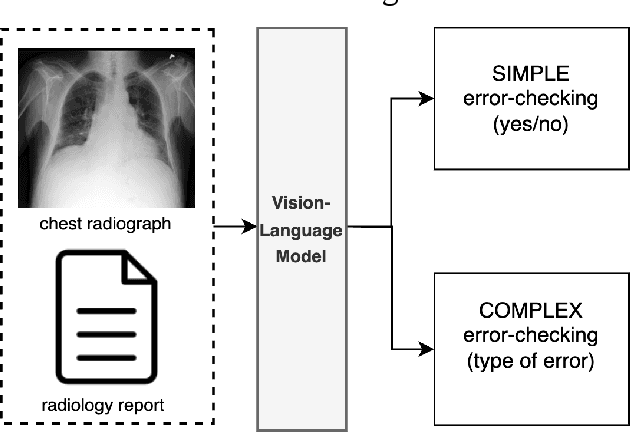
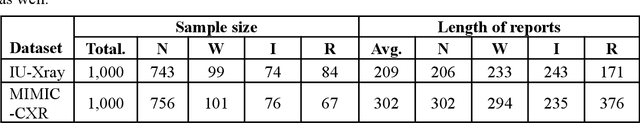

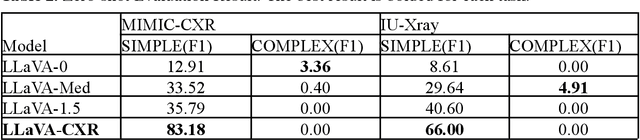
Abstract:This paper proposes one of the first clinical applications of multimodal large language models (LLMs) as an assistant for radiologists to check errors in their reports. We created an evaluation dataset from two real-world radiology datasets (MIMIC-CXR and IU-Xray), with 1,000 subsampled reports each. A subset of original reports was modified to contain synthetic errors by introducing various type of mistakes. The evaluation contained two difficulty levels: SIMPLE for binary error-checking and COMPLEX for identifying error types. LLaVA (Large Language and Visual Assistant) variant models, including our instruction-tuned model, were used for the evaluation. Additionally, a domain expert evaluation was conducted on a small test set. At the SIMPLE level, the LLaVA v1.5 model outperformed other publicly available models. Instruction tuning significantly enhanced performance by 47.4% and 25.4% on MIMIC-CXR and IU-Xray data, respectively. The model also surpassed the domain experts accuracy in the MIMIC-CXR dataset by 1.67%. Notably, among the subsets (N=21) of the test set where a clinician did not achieve the correct conclusion, the LLaVA ensemble mode correctly identified 71.4% of these cases. This study marks a promising step toward utilizing multi-modal LLMs to enhance diagnostic accuracy in radiology. The ensemble model demonstrated comparable performance to clinicians, even capturing errors overlooked by humans. Nevertheless, future work is needed to improve the model ability to identify the types of inconsistency.
Optimising Chest X-Rays for Image Analysis by Identifying and Removing Confounding Factors
Aug 22, 2022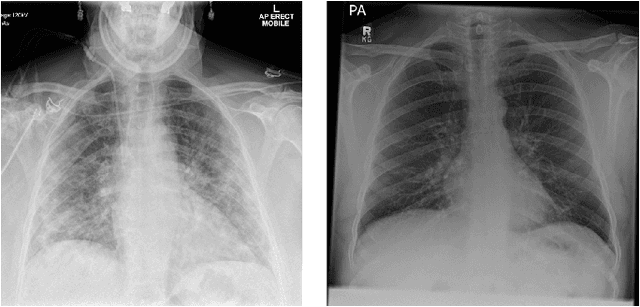
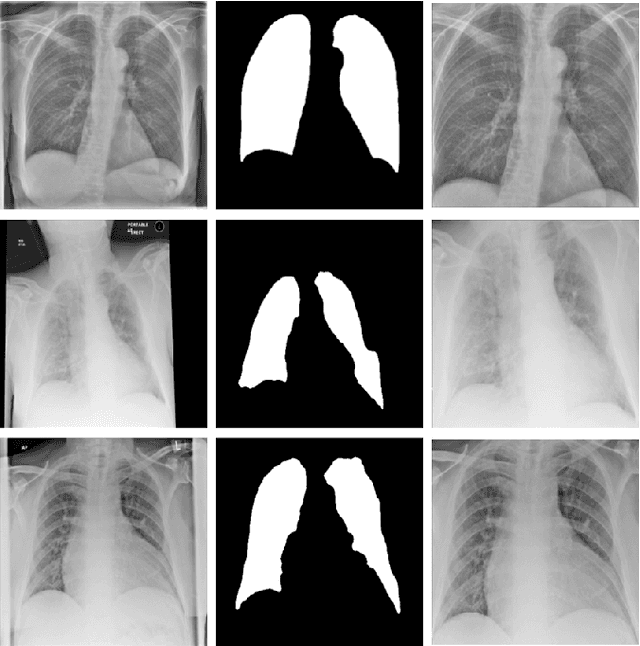
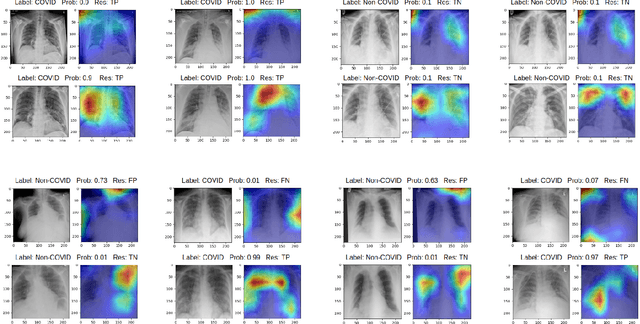
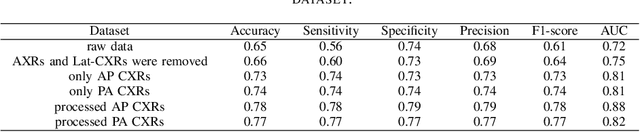
Abstract:During the COVID-19 pandemic, the sheer volume of imaging performed in an emergency setting for COVID-19 diagnosis has resulted in a wide variability of clinical CXR acquisitions. This variation is seen in the CXR projections used, image annotations added and in the inspiratory effort and degree of rotation of clinical images. The image analysis community has attempted to ease the burden on overstretched radiology departments during the pandemic by developing automated COVID-19 diagnostic algorithms, the input for which has been CXR imaging. Large publicly available CXR datasets have been leveraged to improve deep learning algorithms for COVID-19 diagnosis. Yet the variable quality of clinically-acquired CXRs within publicly available datasets could have a profound effect on algorithm performance. COVID-19 diagnosis may be inferred by an algorithm from non-anatomical features on an image such as image labels. These imaging shortcuts may be dataset-specific and limit the generalisability of AI systems. Understanding and correcting key potential biases in CXR images is therefore an essential first step prior to CXR image analysis. In this study, we propose a simple and effective step-wise approach to pre-processing a COVID-19 chest X-ray dataset to remove undesired biases. We perform ablation studies to show the impact of each individual step. The results suggest that using our proposed pipeline could increase accuracy of the baseline COVID-19 detection algorithm by up to 13%.
Dual Encoder Fusion U-Net for Cross-manufacturer Chest X-ray Segmentation
Sep 27, 2020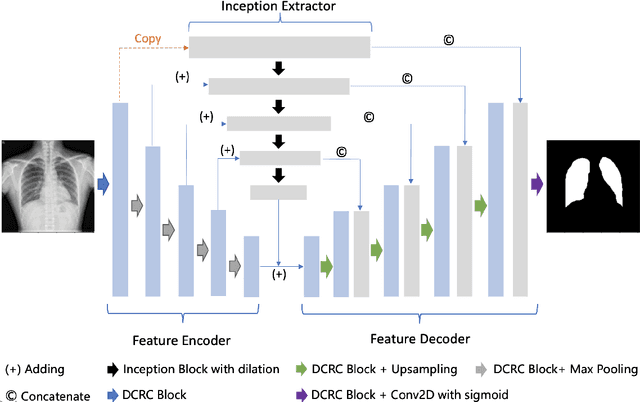
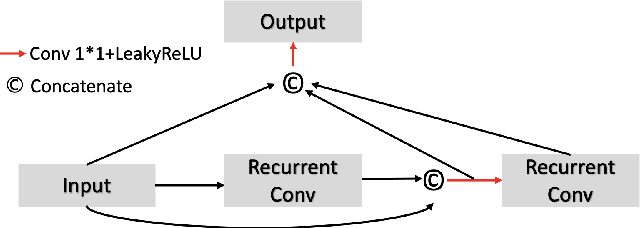
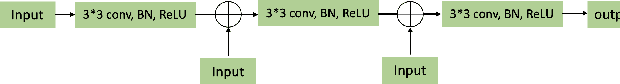
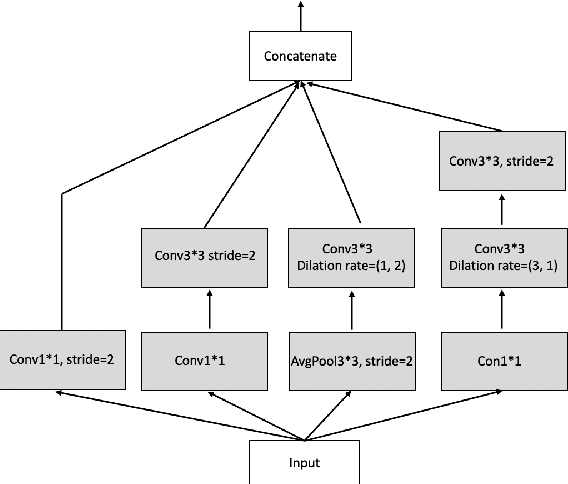
Abstract:A number of methods based on the deep learning have been applied to medical image segmentation and have achieved state-of-the-art performance. Due to the importance of chest x-ray data in studying COVID-19, there is a demand for state-of-the-art models capable of precisely segmenting soft tissue on the chest x-rays before obtaining mask annotations about this sort of dataset. The dataset for exploring best pre-trained model is from Montgomery and Shenzhen hospital which had opened in 2014. The most famous technique is U-Net which has been used to many medical datasets including the Chest X-ray. However, most variant U-Nets mainly focus on extraction of contextual information and skip connection. There is still a large space for improving extraction of spatial features. In this paper, we propose a dual encoder fusion U-Net framework for Chest X-rays based on Inception Convolutional Neural Network with dilation, Densely Connected Recurrent Convolutional Neural Network, which is named DEFU-Net. The densely connected recurrent path extends the network deeper for facilitating context feature extraction. In order to increase the width of network and enrich representation of features, the inception blocks with dilation have been used. The inception blocks can capture globally and locally spatial information by various receptive fields. At the same time, the two paths are fused by summing features, thus preserving context and the spatial information for decoding part. This multi-learning-scale model is benefiting in Chest X-ray dataset from two different manufacturers (Montgomery and Shenzhen hospital). The DEFU-Net achieves the better performance than basic U-Net, residual U-Net, BCDU-Net, modified R2U-Net and modified attention R2U-Net. This model has proved the feasibility for mixed dataset. The open source code for this proposed framework will be public soon.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge