Chengzhe Piao
Privacy Preserved Blood Glucose Level Cross-Prediction: An Asynchronous Decentralized Federated Learning Approach
Jun 21, 2024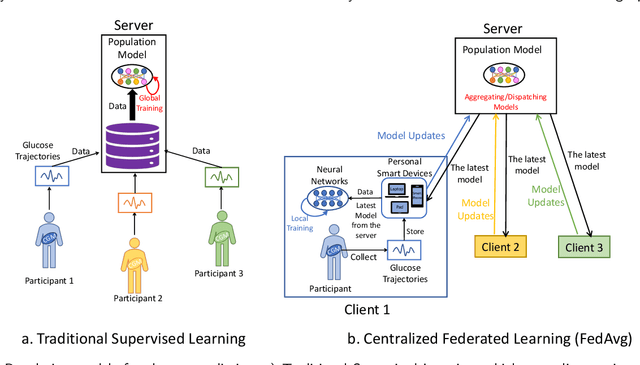
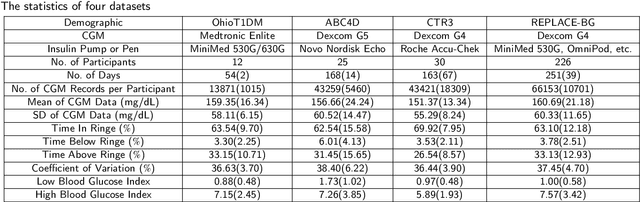
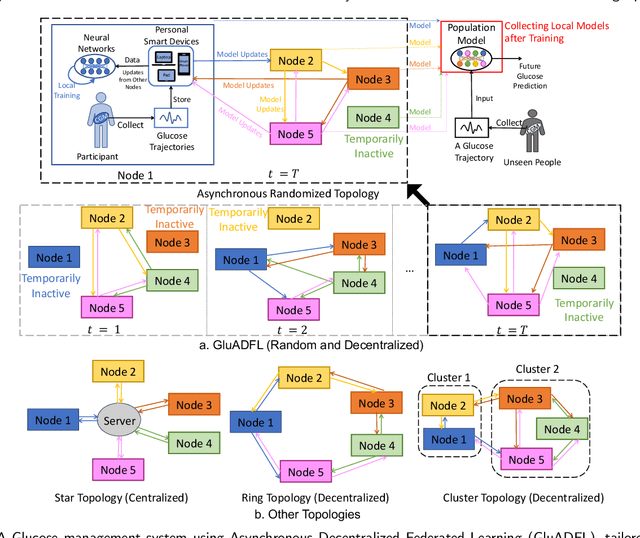
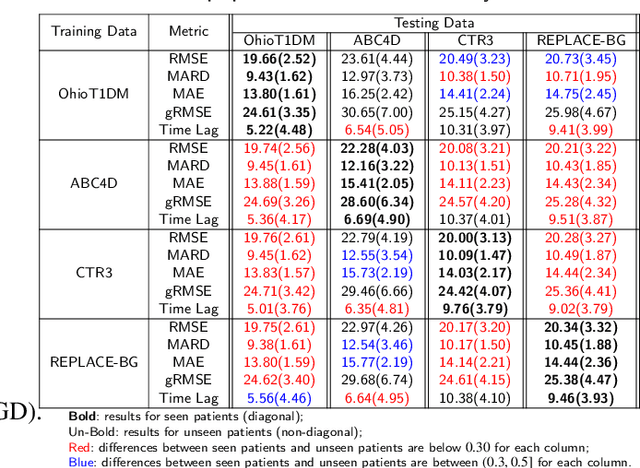
Abstract:Newly diagnosed Type 1 Diabetes (T1D) patients often struggle to obtain effective Blood Glucose (BG) prediction models due to the lack of sufficient BG data from Continuous Glucose Monitoring (CGM), presenting a significant "cold start" problem in patient care. Utilizing population models to address this challenge is a potential solution, but collecting patient data for training population models in a privacy-conscious manner is challenging, especially given that such data is often stored on personal devices. Considering the privacy protection and addressing the "cold start" problem in diabetes care, we propose "GluADFL", blood Glucose prediction by Asynchronous Decentralized Federated Learning. We compared GluADFL with eight baseline methods using four distinct T1D datasets, comprising 298 participants, which demonstrated its superior performance in accurately predicting BG levels for cross-patient analysis. Furthermore, patients' data might be stored and shared across various communication networks in GluADFL, ranging from highly interconnected (e.g., random, performs the best among others) to more structured topologies (e.g., cluster and ring), suitable for various social networks. The asynchronous training framework supports flexible participation. By adjusting the ratios of inactive participants, we found it remains stable if less than 70% are inactive. Our results confirm that GluADFL offers a practical, privacy-preserving solution for BG prediction in T1D, significantly enhancing the quality of diabetes management.
GARNN: An Interpretable Graph Attentive Recurrent Neural Network for Predicting Blood Glucose Levels via Multivariate Time Series
Feb 26, 2024



Abstract:Accurate prediction of future blood glucose (BG) levels can effectively improve BG management for people living with diabetes, thereby reducing complications and improving quality of life. The state of the art of BG prediction has been achieved by leveraging advanced deep learning methods to model multi-modal data, i.e., sensor data and self-reported event data, organised as multi-variate time series (MTS). However, these methods are mostly regarded as ``black boxes'' and not entirely trusted by clinicians and patients. In this paper, we propose interpretable graph attentive recurrent neural networks (GARNNs) to model MTS, explaining variable contributions via summarizing variable importance and generating feature maps by graph attention mechanisms instead of post-hoc analysis. We evaluate GARNNs on four datasets, representing diverse clinical scenarios. Upon comparison with twelve well-established baseline methods, GARNNs not only achieve the best prediction accuracy but also provide high-quality temporal interpretability, in particular for postprandial glucose levels as a result of corresponding meal intake and insulin injection. These findings underline the potential of GARNN as a robust tool for improving diabetes care, bridging the gap between deep learning technology and real-world healthcare solutions.
Blood Glucose Level Prediction: A Graph-based Explainable Method with Federated Learning
Dec 19, 2023Abstract:In the UK, approximately 400,000 people with type 1 diabetes (T1D) rely on insulin delivery due to insufficient pancreatic insulin production. Managing blood glucose (BG) levels is crucial, with continuous glucose monitoring (CGM) playing a key role. CGM, tracking BG every 5 minutes, enables effective blood glucose level prediction (BGLP) by considering factors like carbohydrate intake and insulin delivery. Recent research has focused on developing sequential models for BGLP using historical BG data, incorporating additional attributes such as carbohydrate intake, insulin delivery, and time. These methods have shown notable success in BGLP, with some providing temporal explanations. However, they often lack clear correlations between attributes and their impact on BGLP. Additionally, some methods raise privacy concerns by aggregating participant data to learn population patterns. Addressing these limitations, we introduced a graph attentive memory (GAM) model, combining a graph attention network (GAT) with a gated recurrent unit (GRU). GAT applies graph attention to model attribute correlations, offering transparent, dynamic attribute relationships. Attention weights dynamically gauge attribute significance over time. To ensure privacy, we employed federated learning (FL), facilitating secure population pattern analysis. Our method was validated using the OhioT1DM'18 and OhioT1DM'20 datasets from 12 participants, focusing on 6 key attributes. We demonstrated our model's stability and effectiveness through hyperparameter impact analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge