Daniel C Alexander
Optimising Chest X-Rays for Image Analysis by Identifying and Removing Confounding Factors
Aug 22, 2022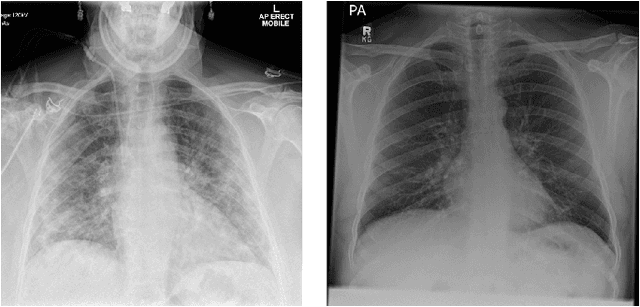
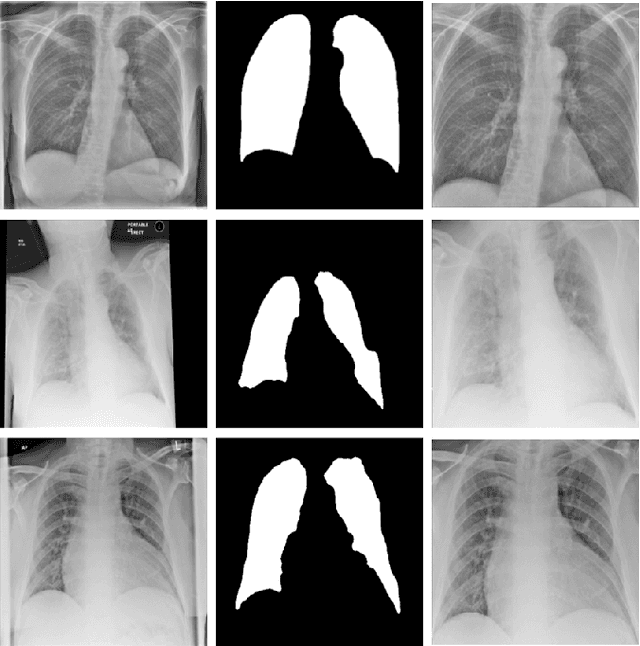
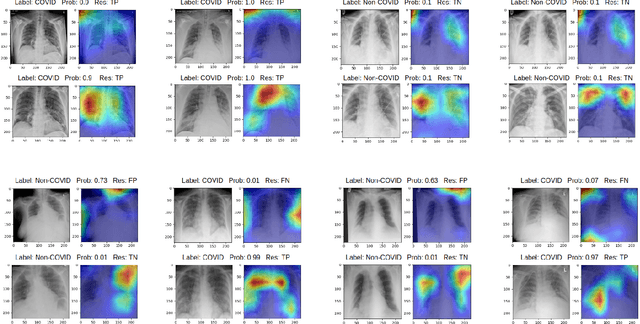
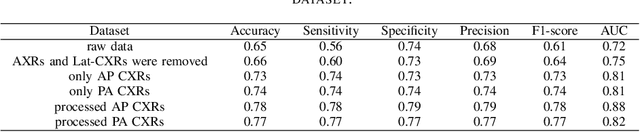
Abstract:During the COVID-19 pandemic, the sheer volume of imaging performed in an emergency setting for COVID-19 diagnosis has resulted in a wide variability of clinical CXR acquisitions. This variation is seen in the CXR projections used, image annotations added and in the inspiratory effort and degree of rotation of clinical images. The image analysis community has attempted to ease the burden on overstretched radiology departments during the pandemic by developing automated COVID-19 diagnostic algorithms, the input for which has been CXR imaging. Large publicly available CXR datasets have been leveraged to improve deep learning algorithms for COVID-19 diagnosis. Yet the variable quality of clinically-acquired CXRs within publicly available datasets could have a profound effect on algorithm performance. COVID-19 diagnosis may be inferred by an algorithm from non-anatomical features on an image such as image labels. These imaging shortcuts may be dataset-specific and limit the generalisability of AI systems. Understanding and correcting key potential biases in CXR images is therefore an essential first step prior to CXR image analysis. In this study, we propose a simple and effective step-wise approach to pre-processing a COVID-19 chest X-ray dataset to remove undesired biases. We perform ablation studies to show the impact of each individual step. The results suggest that using our proposed pipeline could increase accuracy of the baseline COVID-19 detection algorithm by up to 13%.
Enhancing Cancer Prediction in Challenging Screen-Detected Incident Lung Nodules Using Time-Series Deep Learning
Mar 30, 2022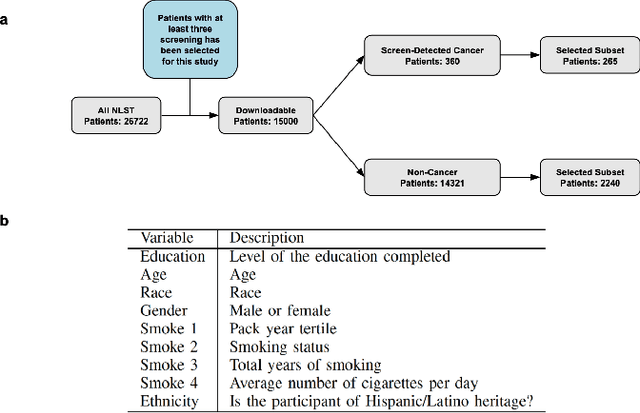
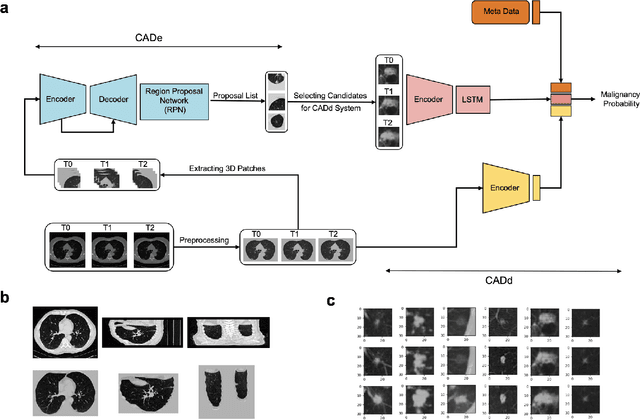
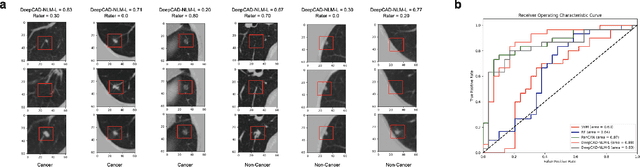

Abstract:Lung cancer is the leading cause of cancer-related mortality worldwide. Lung cancer screening (LCS) using annual low-dose computed tomography (CT) scanning has been proven to significantly reduce lung cancer mortality by detecting cancerous lung nodules at an earlier stage. Improving risk stratification of malignancy risk in lung nodules can be enhanced using machine/deep learning algorithms. However most existing algorithms: a) have primarily assessed single time-point CT data alone thereby failing to utilize the inherent advantages contained within longitudinal imaging datasets; b) have not integrated into computer models pertinent clinical data that might inform risk prediction; c) have not assessed algorithm performance on the spectrum of nodules that are most challenging for radiologists to interpret and where assistance from analytic tools would be most beneficial. Here we show the performance of our time-series deep learning model (DeepCAD-NLM-L) which integrates multi-model information across three longitudinal data domains: nodule-specific, lung-specific, and clinical demographic data. We compared our time-series deep learning model to a) radiologist performance on CTs from the National Lung Screening Trial enriched with the most challenging nodules for diagnosis; b) a nodule management algorithm from a North London LCS study (SUMMIT). Our model demonstrated comparable and complementary performance to radiologists when interpreting challenging lung nodules and showed improved performance (AUC=88\%) against models utilizing single time-point data only. The results emphasise the importance of time-series, multi-modal analysis when interpreting malignancy risk in LCS.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge