Bojidar Rangelov
A data-centric deep learning approach to airway segmentation
Jul 29, 2023Abstract:The morphology and distribution of airway tree abnormalities enables diagnosis and disease characterisation across a variety of chronic respiratory conditions. In this regard, airway segmentation plays a critical role in the production of the outline of the entire airway tree to enable estimation of disease extent and severity. In this study, we propose a data-centric deep learning technique to segment the airway tree. The proposed technique utilises interpolation and image split to improve data usefulness and quality. Then, an ensemble learning strategy is implemented to aggregate the segmented airway trees at different scales. In terms of segmentation performance (dice similarity coefficient), our method outperforms the baseline model by 2.5% on average when a combined loss is used. Further, our proposed technique has a low GPU usage and high flexibility enabling it to be deployed on any 2D deep learning model.
Efficient automatic segmentation for multi-level pulmonary arteries: The PARSE challenge
Apr 07, 2023



Abstract:Efficient automatic segmentation of multi-level (i.e. main and branch) pulmonary arteries (PA) in CTPA images plays a significant role in clinical applications. However, most existing methods concentrate only on main PA or branch PA segmentation separately and ignore segmentation efficiency. Besides, there is no public large-scale dataset focused on PA segmentation, which makes it highly challenging to compare the different methods. To benchmark multi-level PA segmentation algorithms, we organized the first \textbf{P}ulmonary \textbf{AR}tery \textbf{SE}gmentation (PARSE) challenge. On the one hand, we focus on both the main PA and the branch PA segmentation. On the other hand, for better clinical application, we assign the same score weight to segmentation efficiency (mainly running time and GPU memory consumption during inference) while ensuring PA segmentation accuracy. We present a summary of the top algorithms and offer some suggestions for efficient and accurate multi-level PA automatic segmentation. We provide the PARSE challenge as open-access for the community to benchmark future algorithm developments at \url{https://parse2022.grand-challenge.org/Parse2022/}.
Multi-site, Multi-domain Airway Tree Modeling : A Public Benchmark for Pulmonary Airway Segmentation
Mar 10, 2023



Abstract:Open international challenges are becoming the de facto standard for assessing computer vision and image analysis algorithms. In recent years, new methods have extended the reach of pulmonary airway segmentation that is closer to the limit of image resolution. Since EXACT'09 pulmonary airway segmentation, limited effort has been directed to quantitative comparison of newly emerged algorithms driven by the maturity of deep learning based approaches and clinical drive for resolving finer details of distal airways for early intervention of pulmonary diseases. Thus far, public annotated datasets are extremely limited, hindering the development of data-driven methods and detailed performance evaluation of new algorithms. To provide a benchmark for the medical imaging community, we organized the Multi-site, Multi-domain Airway Tree Modeling (ATM'22), which was held as an official challenge event during the MICCAI 2022 conference. ATM'22 provides large-scale CT scans with detailed pulmonary airway annotation, including 500 CT scans (300 for training, 50 for validation, and 150 for testing). The dataset was collected from different sites and it further included a portion of noisy COVID-19 CTs with ground-glass opacity and consolidation. Twenty-three teams participated in the entire phase of the challenge and the algorithms for the top ten teams are reviewed in this paper. Quantitative and qualitative results revealed that deep learning models embedded with the topological continuity enhancement achieved superior performance in general. ATM'22 challenge holds as an open-call design, the training data and the gold standard evaluation are available upon successful registration via its homepage.
Optimising Chest X-Rays for Image Analysis by Identifying and Removing Confounding Factors
Aug 22, 2022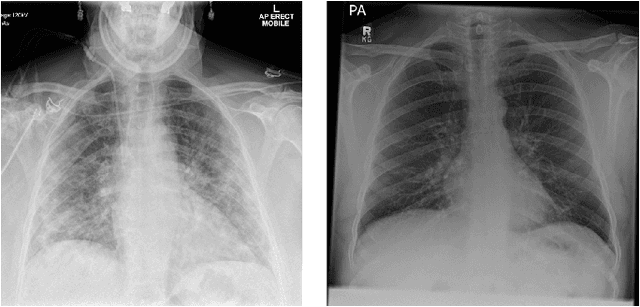
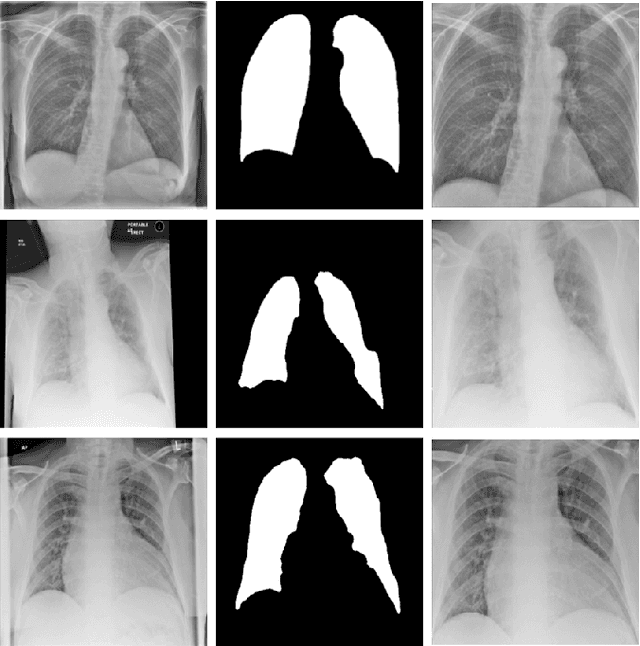
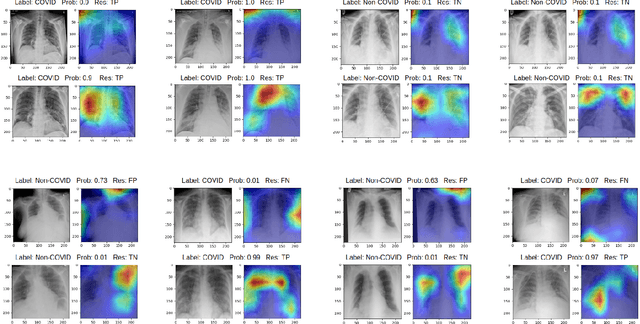
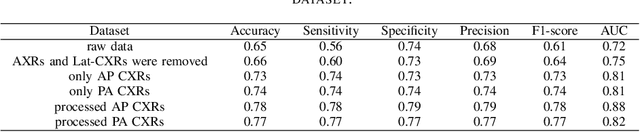
Abstract:During the COVID-19 pandemic, the sheer volume of imaging performed in an emergency setting for COVID-19 diagnosis has resulted in a wide variability of clinical CXR acquisitions. This variation is seen in the CXR projections used, image annotations added and in the inspiratory effort and degree of rotation of clinical images. The image analysis community has attempted to ease the burden on overstretched radiology departments during the pandemic by developing automated COVID-19 diagnostic algorithms, the input for which has been CXR imaging. Large publicly available CXR datasets have been leveraged to improve deep learning algorithms for COVID-19 diagnosis. Yet the variable quality of clinically-acquired CXRs within publicly available datasets could have a profound effect on algorithm performance. COVID-19 diagnosis may be inferred by an algorithm from non-anatomical features on an image such as image labels. These imaging shortcuts may be dataset-specific and limit the generalisability of AI systems. Understanding and correcting key potential biases in CXR images is therefore an essential first step prior to CXR image analysis. In this study, we propose a simple and effective step-wise approach to pre-processing a COVID-19 chest X-ray dataset to remove undesired biases. We perform ablation studies to show the impact of each individual step. The results suggest that using our proposed pipeline could increase accuracy of the baseline COVID-19 detection algorithm by up to 13%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge